Preferential interactions of guanidinum ions with aromatic groups over aliphatic groups
- PMID: 19874022
- PMCID: PMC2784182
- DOI: 10.1021/ja903478s
Preferential interactions of guanidinum ions with aromatic groups over aliphatic groups
Abstract
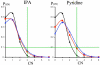
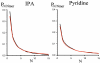
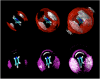
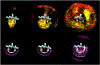
Small angle neutron scattering (SANS) and molecular dynamics (MD) simulations were used to characterize the long-range structuring (aggregation) of aqueous solutions of isopropanol (IPA) and pyridine and the effect on structuring of guanidinium chloride (GdmCl). These solutes serve as highly soluble analogs of the nonpolar aliphatic (IPA) and aromatic (pyridine) side chains of proteins. SANS data showed that isopropanol and pyridine both form clusters in water resulting from interaction between nonpolar groups of the solutes, with pyridine aggregation producing longer-range structuring than isopropanol in 3 m solutions. Addition of GdmCl at 3 m concentration considerably reduced pyridine aggregation but had no effect on isopropanol aggregation. MD simulations of these solutions support the conclusion that long-range structuring involves hydrophobic solute interactions and that Gdm(+) interacts with the planar pyridine group to suppress pyridine-pyridine interactions in solution. Hydrophobic interactions involving the aliphatic groups of isopropanol were unaffected by GdmCl, indicating that the planar and weakly hydrated Gdm(+) cation cannot make productive interactions with the highly curved or "lumpy" aliphatic groups of this solute. These observations support the conclusion that the effects of Gdm(+) ions on protein-stabilizing interactions involving aromatic amino acid side chains make significant contributions to the denaturant activity of GdmCl, whereas interactions with the "lumpy" aliphatic side chains are likely to be less important.
Figures



Similar articles
-
The structure of aqueous guanidinium chloride solutions.J Am Chem Soc. 2004 Sep 22;126(37):11462-70. doi: 10.1021/ja040034x. J Am Chem Soc. 2004. PMID: 15366892
-
Effects of Concentration on Like-Charge Pairing of Guanidinium Ions and on the Structure of Water: An All-Atom Molecular Dynamics Simulation Study.J Phys Chem B. 2015 Aug 27;119(34):11262-74. doi: 10.1021/acs.jpcb.5b03064. Epub 2015 Jul 20. J Phys Chem B. 2015. PMID: 26132632
-
Unfolding of hydrophobic polymers in guanidinium chloride solutions.J Phys Chem B. 2010 Feb 18;114(6):2246-54. doi: 10.1021/jp906976q. J Phys Chem B. 2010. PMID: 20146543
-
Suppression of protein interactions by arginine: a proposed mechanism of the arginine effects.Biophys Chem. 2007 Apr;127(1-2):1-8. doi: 10.1016/j.bpc.2006.12.007. Epub 2006 Dec 21. Biophys Chem. 2007. PMID: 17257734 Review.
-
Weakly hydrated surfaces and the binding interactions of small biological solutes.Eur Biophys J. 2012 Apr;41(4):369-77. doi: 10.1007/s00249-011-0776-2. Epub 2011 Nov 29. Eur Biophys J. 2012. PMID: 22124617 Review.
Cited by
-
A new structural technique for examining ion-neutral association in aqueous solution.Faraday Discuss. 2013;160:161-70; discussion 207-24. doi: 10.1039/c2fd20081c. Faraday Discuss. 2013. PMID: 23795499 Free PMC article.
-
Effect of stepwise microhydration on the methylammonium···phenol and ammonium···phenol interaction.J Mol Model. 2013 May;19(5):1985-94. doi: 10.1007/s00894-012-1579-9. Epub 2012 Sep 8. J Mol Model. 2013. PMID: 22961590
-
Hydration of guanidinium depends on its local environment.Chem Sci. 2015 Jun 1;6(6):3420-3429. doi: 10.1039/c5sc00618j. Epub 2015 Apr 14. Chem Sci. 2015. PMID: 28706704 Free PMC article.
-
Quantitative assessments of the distinct contributions of polypeptide backbone amides versus side chain groups to chain expansion via chemical denaturation.J Am Chem Soc. 2015 Mar 4;137(8):2984-95. doi: 10.1021/ja512062h. Epub 2015 Feb 23. J Am Chem Soc. 2015. PMID: 25664638 Free PMC article.
-
Orientation of Methylguanidinium Ions at the Water-Air Interface.J Phys Chem C Nanomater Interfaces. 2017 Oct 26;121(42):23398-23405. doi: 10.1021/acs.jpcc.7b03752. Epub 2017 Sep 14. J Phys Chem C Nanomater Interfaces. 2017. PMID: 29129985 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

