SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice
- PMID: 19874786
- PMCID: PMC2788510
- DOI: 10.1016/j.neuron.2009.08.018
SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice
Abstract
Nuclear movement is critical during neurogenesis and neuronal migration, which are fundamental for mammalian brain development. Although dynein, Lis1, and other cytoplasmic proteins are known for their roles in connecting microtubules to the nucleus during interkinetic nuclear migration (INM) and nucleokinesis, the factors connecting dynein/Lis1 to the nuclear envelope (NE) remain to be determined. We report here that the SUN-domain proteins SUN1 and SUN2 and the KASH-domain proteins Syne-1/Nesprin-1 and Syne-2/Nesprin-2 play critical roles in neurogenesis and neuronal migration in mice. We show that SUN1 and SUN2 redundantly form complexes with Syne-2 to mediate the centrosome-nucleus coupling during both INM and radial neuronal migration in the cerebral cortex. Syne-2 is connected to the centrosome through interactions with both dynein/dynactin and kinesin complexes. Syne-2 mutants also display severe defects in learning and memory. These results fill an important gap in our understanding of the mechanism of nuclear movement during brain development.
Figures

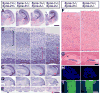
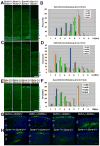


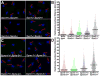

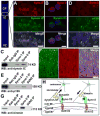
Comment in
-
Sun proteins enlighten nuclear movement in development.Neuron. 2009 Oct 29;64(2):147-9. doi: 10.1016/j.neuron.2009.10.010. Neuron. 2009. PMID: 19874779 Free PMC article.
References
-
- Chae T, Kwon YT, Bronson R, Dikkes P, Li E, Tsai LH. Mice lacking p35, a neuronal specific activator of Cdk5, display cortical lamination defects, seizures, and adult lethality. Neuron. 1997;18:29–42. - PubMed
-
- Chen G, Sima J, Jin M, Wang KY, Xue XJ, Zheng W, Ding YQ, Yuan XB. Semaphorin-3A guides radial migration of cortical neurons during development. Nat Neurosci. 2008;11:36–44. - PubMed
-
- D'Arcangelo G, Miao GG, Chen SC, Soares HD, Morgan JI, Curran T. A protein related to extracellular matrix proteins deleted in the mouse mutant reeler. Nature. 1995;374:719–723. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

