Directed evolution and structural characterization of a simvastatin synthase
- PMID: 19875080
- PMCID: PMC2798062
- DOI: 10.1016/j.chembiol.2009.09.017
Directed evolution and structural characterization of a simvastatin synthase
Abstract
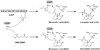
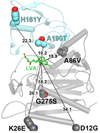
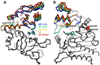
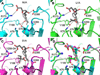
Enzymes from natural product biosynthetic pathways are attractive candidates for creating tailored biocatalysts to produce semisynthetic pharmaceutical compounds. LovD is an acyltransferase that converts the inactive monacolin J acid (MJA) into the cholesterol-lowering lovastatin. LovD can also synthesize the blockbuster drug simvastatin using MJA and a synthetic alpha-dimethylbutyryl thioester, albeit with suboptimal properties as a biocatalyst. Here we used directed evolution to improve the properties of LovD toward semisynthesis of simvastatin. Mutants with improved catalytic efficiency, solubility, and thermal stability were obtained, with the best mutant displaying an approximately 11-fold increase in an Escherichia coli-based biocatalytic platform. To understand the structural basis of LovD enzymology, seven X-ray crystal structures were determined, including the parent LovD, an improved mutant G5, and G5 cocrystallized with ligands. Comparisons between the structures reveal that beneficial mutations stabilize the structure of G5 in a more compact conformation that is favorable for catalysis.
Figures


References
-
- Arnold FH. Combinatorial and computational challenges for biocatalyst design. Nature. 2001;409:253–257. - PubMed
-
- Arnold FH, Volkov AA. Directed evolution of biocatalysts. Curr. Opin. Chem. Biol. 1999;3:54–59. - PubMed
-
- Berg VA, Hans M, Steekstra H. Method for the production of simvastatin. 2009 WO 2007147801 (A1)
-
- Fromant M, Blanquet S, Plateau P. Direct random mutagenesis of gene-sized DNA fragments using polymerase chain-reaction. Anal. Biochem. 1995;224:347–353. - PubMed
-
- Heikinheimo P, Goldman A, Jeffries C, Ollis DL. Of barn owls and bankers: a lush variety of alpha/beta hydrolases. Structure. 1999;7:141–146. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

