Can literature analysis identify innovation drivers in drug discovery?
- PMID: 19876041
- PMCID: PMC7097144
- DOI: 10.1038/nrd2973
Can literature analysis identify innovation drivers in drug discovery?
Abstract
Drug discovery must be guided not only by medical need and commercial potential, but also by the areas in which new science is creating therapeutic opportunities, such as target identification and the understanding of disease mechanisms. To systematically identify such areas of high scientific activity, we use bibliometrics and related data-mining methods to analyse over half a terabyte of data, including PubMed abstracts, literature citation data and patent filings. These analyses reveal trends in scientific activity related to disease studied at varying levels, down to individual genes and pathways, and provide methods to monitor areas in which scientific advances are likely to create new therapeutic opportunities.
Figures
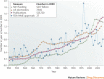


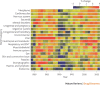
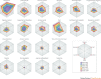
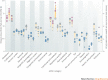

Similar articles
-
What makes a good drug target?Drug Discov Today. 2011 Dec;16(23-24):1037-43. doi: 10.1016/j.drudis.2011.09.007. Epub 2011 Sep 16. Drug Discov Today. 2011. PMID: 21945861 Review.
-
The need for scientific software engineering in the pharmaceutical industry.J Comput Aided Mol Des. 2017 Mar;31(3):301-304. doi: 10.1007/s10822-016-9997-x. Epub 2016 Dec 19. J Comput Aided Mol Des. 2017. PMID: 27995514
-
The impact of chemoinformatics on drug discovery in the pharmaceutical industry.Expert Opin Drug Discov. 2020 Mar;15(3):293-306. doi: 10.1080/17460441.2020.1696307. Epub 2020 Jan 22. Expert Opin Drug Discov. 2020. PMID: 31965870
-
Mining emerging biomedical literature for understanding disease associations in drug discovery.Methods Mol Biol. 2014;1159:171-206. doi: 10.1007/978-1-4939-0709-0_11. Methods Mol Biol. 2014. PMID: 24788268 Review.
-
Society for Biomolecular Sciences--15th Annual Conference. Bioassay and technology innovation: 15 years of shaping drug discovery. 26-30 April 2009, Lille, France.IDrugs. 2009 Jun;12(6):363-5. IDrugs. 2009. PMID: 19517316 No abstract available.
Cited by
-
Patterns of technological innovation in biotech.Nat Biotechnol. 2012 Oct;30(10):937-43. doi: 10.1038/nbt.2389. Nat Biotechnol. 2012. PMID: 23051809 Review. No abstract available.
-
Adoption of the Citation Typing Ontology by the Journal of Cheminformatics.J Cheminform. 2020 Aug 6;12(1):47. doi: 10.1186/s13321-020-00448-1. J Cheminform. 2020. PMID: 33430994 Free PMC article. No abstract available.
-
Empowering industrial research with shared biomedical vocabularies.Drug Discov Today. 2011 Nov;16(21-22):940-7. doi: 10.1016/j.drudis.2011.09.013. Epub 2011 Sep 23. Drug Discov Today. 2011. PMID: 21963522 Free PMC article. Review.
-
Tracking 20 years of compound-to-target output from literature and patents.PLoS One. 2013 Oct 29;8(10):e77142. doi: 10.1371/journal.pone.0077142. eCollection 2013. PLoS One. 2013. PMID: 24204758 Free PMC article.
-
Quantitative biomedical annotation using medical subject heading over-representation profiles (MeSHOPs).BMC Bioinformatics. 2012 Sep 27;13:249. doi: 10.1186/1471-2105-13-249. BMC Bioinformatics. 2012. PMID: 23017167 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

