Acquisition of aneuploidy provides increased fitness during the evolution of antifungal drug resistance
- PMID: 19876375
- PMCID: PMC2760147
- DOI: 10.1371/journal.pgen.1000705
Acquisition of aneuploidy provides increased fitness during the evolution of antifungal drug resistance
Abstract
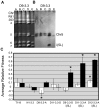
The evolution of drug resistance is an important process that affects clinical outcomes. Resistance to fluconazole, the most widely used antifungal, is often associated with acquired aneuploidy. Here we provide a longitudinal study of the prevalence and dynamics of gross chromosomal rearrangements, including aneuploidy, in the presence and absence of fluconazole during a well-controlled in vitro evolution experiment using Candida albicans, the most prevalent human fungal pathogen. While no aneuploidy was detected in any of the no-drug control populations, in all fluconazole-treated populations analyzed an isochromosome 5L [i(5L)] appeared soon after drug exposure. This isochromosome was associated with increased fitness in the presence of drug and, over time, became fixed in independent populations. In two separate cases, larger supernumerary chromosomes composed of i(5L) attached to an intact chromosome or chromosome fragment formed during exposure to the drug. Other aneuploidies, particularly trisomies of the smaller chromosomes (Chr3-7), appeared throughout the evolution experiment, and the accumulation of multiple aneuploid chromosomes per cell coincided with the highest resistance to fluconazole. Unlike the case in many other organisms, some isolates carrying i(5L) exhibited improved fitness in the presence, as well as in the absence, of fluconazole. The early appearance of aneuploidy is consistent with a model in which C. albicans becomes more permissive of chromosome rearrangements and segregation defects in the presence of fluconazole.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







References
-
- Cannon RD, Lamping E, Holmes AR, Niimi K, Tanabe K, et al. Candida albicans drug resistance another way to cope with stress. Microbiology. 2007;153:3211–3217. - PubMed
-
- Cowen LE. The evolution of fungal drug resistance: modulating the trajectory from genotype to phenotype. Nat Rev Microbiol. 2008;6:187–198. - PubMed
-
- Marr KA, Seidel K, White TC, Bowden RA. Candidemia in allogeneic blood and marrow transplant recipients: evolution of risk factors after the adoption of prophylactic fluconazole. J Infect Dis. 2000;181:309–316. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

