Micro-RNA-155 inhibits IFN-gamma signaling in CD4+ T cells
- PMID: 19877012
- PMCID: PMC2807623
- DOI: 10.1002/eji.200939381
Micro-RNA-155 inhibits IFN-gamma signaling in CD4+ T cells
Abstract
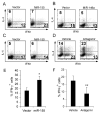
Micro-RNA (miR) are increasingly recognized as critical regulators of tissue-specific patterns of gene expression. CD4+ T cells lacking miR-155, for example, exhibit bias towards Th2 differentiation, indicating that the absence of individual miR could alter CD4+ T-cell differentiation. We now show that miR-155 is induced upon T-cell activation and that it promotes Th1 differentiation when over-expressed in activated CD4+ T cells. Antagonism of miR-155 leads to induction of IFN-gamma receptor alpha-chain (IFN-gammaRalpha), and a functional miR-155 target site is identified within the 3' untranslated region of IFN-gammaRalpha. These results identify IFN-gammaRalpha as a second miR-155 target in T cells and suggest that miR-155 contributes to Th1 differentiation in CD4+ T cells by inhibiting IFN-gamma signaling.
Conflict of interest statement
Conflict of interest: The authors declare no financial or commercial conflict of interest.
Figures


References
-
- Reiner SL. Development in motion: helper T cells at work. Cell. 2007;129:33–36. - PubMed
-
- Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P, Klein LO, Davis MM, Chen CZ. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. - PubMed
-
- Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

