Phylogenetic analysis of mitochondrial substitution rate variation in the angiosperm tribe Sileneae
- PMID: 19878576
- PMCID: PMC2777880
- DOI: 10.1186/1471-2148-9-260
Phylogenetic analysis of mitochondrial substitution rate variation in the angiosperm tribe Sileneae
Erratum in
- BMC Evol Biol. 2010;10:12
Abstract
Background: Recent phylogenetic studies have revealed that the mitochondrial genome of the angiosperm Silene noctiflora (Caryophyllaceae) has experienced a massive mutation-driven acceleration in substitution rate, placing it among the fastest evolving eukaryotic genomes ever identified. To date, it appears that other species within Silene have maintained more typical substitution rates, suggesting that the acceleration in S. noctiflora is a recent and isolated evolutionary event. This assessment, however, is based on a very limited sampling of taxa within this diverse genus.
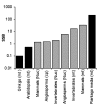
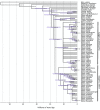
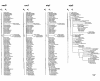
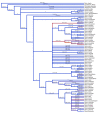
Results: We analyzed the substitution rates in 4 mitochondrial genes (atp1, atp9, cox3 and nad9) across a broad sample of 74 species within Silene and related genera in the tribe Sileneae. We found that S. noctiflora shares its history of elevated mitochondrial substitution rate with the closely related species S. turkestanica. Another section of the genus (Conoimorpha) has experienced an acceleration of comparable magnitude. The phylogenetic data remain ambiguous as to whether the accelerations in these two clades represent independent evolutionary events or a single ancestral change. Rate variation among genes was equally dramatic. Most of the genus exhibited elevated rates for atp9 such that the average tree-wide substitution rate for this gene approached the values for the fastest evolving branches in the other three genes. In addition, some species exhibited major accelerations in atp1 and/or cox3 with no correlated change in other genes. Rates of non-synonymous substitution did not increase proportionally with synonymous rates but instead remained low and relatively invariant.
Conclusion: The patterns of phylogenetic divergence within Sileneae suggest enormous variability in plant mitochondrial mutation rates and reveal a complex interaction of gene and species effects. The variation in rates across genomic and phylogenetic scales raises questions about the mechanisms responsible for the evolution of mutation rates in plant mitochondrial genomes.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

