Phylogenetic relatedness and host plant growth form influence gene expression of the polyphagous comma butterfly (Polygonia c-album)
- PMID: 19878603
- PMCID: PMC2775755
- DOI: 10.1186/1471-2164-10-506
Phylogenetic relatedness and host plant growth form influence gene expression of the polyphagous comma butterfly (Polygonia c-album)
Abstract
Background: The mechanisms that shape the host plant range of herbivorous insect are to date not well understood but knowledge of these mechanisms and the selective forces that influence them can expand our understanding of the larger ecological interaction. Nevertheless, it is well established that chemical defenses of plants influence the host range of herbivorous insects. While host plant chemistry is influenced by phylogeny, also the growth forms of plants appear to influence the plant defense strategies as first postulated by Feeny (the "plant apparency" hypothesis). In the present study we aim to investigate the molecular basis of the diverse host plant range of the comma butterfly (Polygonia c-album) by testing differential gene expression in the caterpillars on three host plants that are either closely related or share the same growth form.
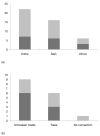
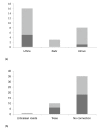
Results: In total 120 genes were identified to be differentially expressed in P. c-album after feeding on different host plants, 55 of them in the midgut and 65 in the restbody of the caterpillars. Expression patterns could be confirmed with an independent method for 14 of 27 tested genes. Pairwise similarities in upregulation in the midgut of the caterpillars were higher between plants that shared either growth form or were phylogenetically related. No known detoxifying enzymes were found to be differently regulated in the midgut after feeding on different host plants.
Conclusion: Our data suggest a complex picture of gene expression in response to host plant feeding. While each plant requires a unique gene regulation in the caterpillar, both phylogenetic relatedness and host plant growth form appear to influence the expression profile of the polyphagous comma butterfly, in agreement with phylogenetic studies of host plant utilization in butterflies.
Figures

References
-
- Dethier VG. Chemical factors determining the choice of food plants by Papilio larvae. Am Nat. 1941;75:61–73.
-
- Ehrlich PR, Raven PH. Butterflies and plants: a study in coevoution. Evolution. 1964;18:586–608.
-
- Fraenkel GS. The raison d'être of secondary plant substances. Science. 1959;129:1466–1470. - PubMed
-
- Thorsteinson AJ. Host selection in phytophagous insects. Annu Rev Entomol. 1960;5:193–218.
-
- Bernays EA. Host range in phytophagous insects - the potential role of generalist predators. Evol Ecol. 1989;3:299–311.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

