PepX: a structural database of non-redundant protein-peptide complexes
- PMID: 19880386
- PMCID: PMC2808939
- DOI: 10.1093/nar/gkp893
PepX: a structural database of non-redundant protein-peptide complexes
Abstract
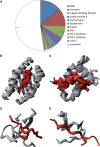
Although protein-peptide interactions are estimated to constitute up to 40% of all protein interactions, relatively little information is available for the structural details of these interactions. Peptide-mediated interactions are a prime target for drug design because they are predominantly present in signaling and regulatory networks. A reliable data set of nonredundant protein-peptide complexes is indispensable as a basis for modeling and design, but current data sets for protein-peptide interactions are often biased towards specific types of interactions or are limited to interactions with small ligands. In PepX (http://pepx.switchlab.org), we have designed an unbiased and exhaustive data set of all protein-peptide complexes available in the Protein Data Bank with peptide lengths up to 35 residues. In addition, these complexes have been clustered based on their binding interfaces rather than sequence homology, providing a set of structurally diverse protein-peptide interactions. The final data set contains 505 unique protein-peptide interface clusters from 1431 complexes. Thorough annotation of each complex with both biological and structural information facilitates searching for and browsing through individual complexes and clusters. Moreover, we provide an additional source of data for peptide design by annotating peptides with naturally occurring backbone variations using fragment clusters from the BriX database.
Figures

References
-
- Neduva V, Russell RB. Peptides mediating interaction networks: new leads at last. Curr. Opin. Biotechnol. 2006;17:465–471. - PubMed
-
- Petsalaki E, Russell RB. Peptide-mediated interactions in biological systems: new discoveries and applications. Curr. Opin. Biotechnol. 2008;19:344–350. - PubMed
-
- Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

