DNA structure specificity conferred on a replicative helicase by its loader
- PMID: 19880515
- PMCID: PMC2801299
- DOI: 10.1074/jbc.M109.072520
DNA structure specificity conferred on a replicative helicase by its loader
Abstract
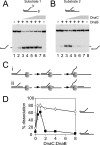
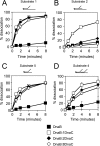
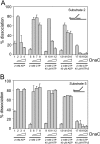
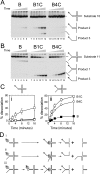
Prokaryotic and eukaryotic replicative helicases can translocate along single-stranded and double-stranded DNA, with the central cavity of these multimeric ring helicases being able to accommodate both forms of DNA. Translocation by such helicases along single-stranded DNA results in the unwinding of forked DNA by steric exclusion and appears critical in unwinding of parental strands at the replication fork, whereas translocation over double-stranded DNA has no well-defined role. We have found that the accessory factor, DnaC, that promotes loading of the Escherichia coli replicative helicase DnaB onto single-stranded DNA may also act to confer DNA structure specificity on DnaB helicase. When present in excess, DnaC inhibits DnaB translocation over double-stranded DNA but not over single-stranded DNA. Inhibition of DnaB translocation over double-stranded DNA requires the ATP-bound form of DnaC, and this inhibition is relieved during translocation over single-stranded DNA indicating that stimulation of DnaC ATPase is responsible for this DNA structure specificity. These findings demonstrate that DnaC may provide the DNA structure specificity lacking in DnaB, limiting DnaB translocation to bona fide replication forks. The ability of other replicative helicases to translocate along single-stranded and double-stranded DNA raises the possibility that analogous regulatory mechanisms exist in other organisms.
Figures


References
-
- Singleton M. R., Dillingham M. S., Wigley D. B. (2007) Annu. Rev. Biochem. 76, 23–50 - PubMed
-
- Patel S. S., Picha K. M. (2000) Annu. Rev. Biochem. 69, 651–697 - PubMed
-
- Kim S., Dallmann H. G., McHenry C. S., Marians K. J. (1996) Cell 84, 643–650 - PubMed
-
- LeBowitz J. H., McMacken R. (1986) J. Biol. Chem. 261, 4738–4748 - PubMed
-
- Baker T. A., Funnell B. E., Kornberg A. (1987) J. Biol. Chem. 262, 6877–6885 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

