A novel distal enhancer mediates cytokine induction of mouse RANKl gene expression
- PMID: 19880655
- PMCID: PMC2796147
- DOI: 10.1210/me.2009-0209
A novel distal enhancer mediates cytokine induction of mouse RANKl gene expression
Abstract
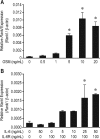
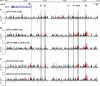
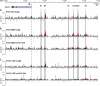
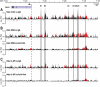
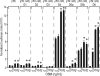
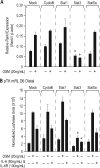
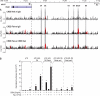
Chronic inflammatory states are associated with increased bone loss. This increase is often linked to an elevation in receptor activator of nuclear factor-kappaB ligand (RANKL), a TNFalpha-like factor essential to osteoclast formation. In this study, we document the ability of IL-6 in combination with IL-6 soluble receptor (IL-6/IL-6sR) and oncostatin M to induce Rankl expression in stromal cells via signal transducer and activator of transcription 3 (STAT3). We used chromatin immunoprecipitation-tiled DNA microarray analysis to determine sites of action of STAT3 at the Rankl locus and to assess the consequences of binding on histone H4 acetylation and RNA polymerase II recruitment. Both IL-6/IL-6 soluble receptor and oncostatin M stimulated STAT3 binding upstream of the Rankl transcriptional start site. Although previously identified enhancers bound STAT3, a more distal enhancer termed mRLD6 was a particular focus of STAT3 binding. When fused to a heterologous promoter, this enhancer was highly active, containing two functionally active STAT response elements. Importantly, small interfering RNA knockdown of Stat3 mRNA and protein, but not that of Stat1 or Stat5a, was effective in limiting Rankl mRNA up-regulation. Interestingly, although RNA polymerase II and histone H4 acetylation marked many of the enhancers under basal conditions, the levels of both were strongly increased after cytokine treatment, particularly at mRLD6. Finally, mRLD6 was also a target for forskolin-induced cellular response element-binding protein (CREB) recruitment, which potentiated cytokine activity. Our studies provide new insight into mechanisms by which glycoprotein 130 activating cytokines induce RANKL expression.
Figures


References
-
- Suda T, Takahashi N, Martin TJ 1992 Modulation of osteoclast differentiation. Endocr Rev 13:66–80 - PubMed
-
- Teitelbaum SL 2000 Bone resorption by osteoclasts. Science 289:1504–1508 - PubMed
-
- Jimi E, Akiyama S, Tsurukai T, Okahashi N, Kobayashi K, Udagawa N, Nishihara T, Takahashi N, Suda T 1999 Osteoclast differentiation factor acts as a multifunctional regulator in murine osteoclast differentiation and function. J Immunol 163:434–442 - PubMed
-
- Lacey DL, Timms E, Tan HL, Kelley MJ, Dunstan CR, Burgess T, Elliott R, Colombero A, Elliott G, Scully S, Hsu H, Sullivan J, Hawkins N, Davy E, Capparelli C, Eli A, Qian YX, Kaufman S, Sarosi I, Shalhoub V, Senaldi G, Guo J, Delaney J, Boyle WJ 1998 Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell 93:165–176 - PubMed
-
- Bernstein CN, Leslie WD 2003 The pathophysiology of bone disease in gastrointestinal disease. Eur J Gastroenterol Hepatol 15:857–864 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

