Loss of parity between IL-2 and IL-21 in the NOD Idd3 locus
- PMID: 19880748
- PMCID: PMC2780811
- DOI: 10.1073/pnas.0903561106
Loss of parity between IL-2 and IL-21 in the NOD Idd3 locus
Abstract
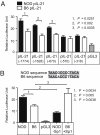
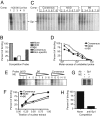
IL-2 and IL-21 are two cytokines with great potential to affect autoimmune infiltration of nonlymphoid tissue, and are contained within the strongest non-MHC-linked locus for type 1 diabetes (T1D) susceptibility on the nonobese diabetic (NOD) mouse (Idd3). IL-21 is necessary for the development of diabetes in the NOD mouse, but a number of important studies argue that decreased expression of IL-2 explains Idd3. In this study, we demonstrate that the amount of IL-21, but not IL-2, correlated with T1D incidence. During our analyses of the IL-2/IL-21 interval, we found that mice segregate into one of two distinct expression profiles. In the first group, which includes the C57BL/6 strain, both Il2 and Il21 were expressed at low levels. In the other group, which includes the NOD strain, Il2 and Il21 were both highly expressed. However, because NOD IL-2 mRNA was relatively unstable, IL-2 production was remarkably similar between strains. The increased production of IL-21 in NOD mice was found to result from two single nucleotide polymorphisms within the distal promoter region that conferred increased binding affinity for the transcription factor Sp1. Our findings indicate that a loss of locus parity after decreased IL-2 mRNA stability ensures that the high-expressing IL-21 allele persists in nature and provides a basis for autoimmunity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Denny P, et al. Mapping of the IDDM locus Idd3 to a 0.35-cM interval containing the interleukin-2 gene. Diabetes. 1997;46:695–700. - PubMed
-
- Todd JA, Wicker LS. Genetic protection from the inflammatory disease type 1 diabetes in humans and animal models. Immunity. 2001;15:387–395. - PubMed
-
- Allison J, et al. Genetic requirements for acceleration of diabetes in non-obese diabetic mice expressing interleukin-2 in islet beta-cells. Eur J Immunol. 1994;24:2535–2541. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

