A zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice
- PMID: 19880795
- PMCID: PMC2782276
- DOI: 10.1105/tpc.109.070771
A zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice
Abstract
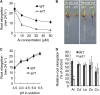
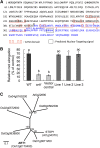
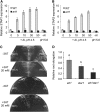
Aluminum (Al) toxicity is the major limiting factor of crop production on acid soils, but some plant species have evolved ways of detoxifying Al. Here, we report a C2H2-type zinc finger transcription factor ART1 (for Al resistance transcription factor 1), which specifically regulates the expression of genes related to Al tolerance in rice (Oryza sativa). ART1 is constitutively expressed in the root, and the expression level is not affected by Al treatment. ART1 is localized in the nucleus of all root cells. A yeast one-hybrid assay showed that ART1 has a transcriptional activation potential and interacts with the promoter region of STAR1, an important factor in rice Al tolerance. Microarray analysis revealed 31 downstream transcripts regulated by ART1, including STAR1 and 2 and a couple of homologs of Al tolerance genes in other plants. Some of these genes were implicated in both internal and external detoxification of Al at different cellular levels. Our findings shed light on comprehensively understanding how plants detoxify aluminum to survive in an acidic environment.
Figures


References
-
- Barceló, J., and Poschenrieder, C. (2002). Fast root growth responses, root exudates, and internal detoxification as clues to the mechanisms of aluminium toxicity and resistance: A review. Environ. Exp. Bot. 48 75–92.
-
- Deng, W., Luo, K., Li, D., Zheng, X., Wei, X., Smith, W., Thammina, C., Lu, L., Li, Y., and Pei, Y. (2006). Overexpression of an Arabidopsis magnesium transport gene, AtMGT1, in Nicotiana benthamiana confers Al tolerance. J. Exp. Bot. 57 4235–4243. - PubMed
-
- Foy, C.D. (1988). Plant adaptation to acid, aluminum-toxic soils. Commun. Soil Sci. Plant Anal. 19 959–987.
-
- Furukawa, J., Yamaji, N., Wang, H., Mitani, N., Murata, Y., Sato, K., Katsuhara, M., Takeda, K., and Ma, J.F. (2007). An aluminum-activated citrate transporter in barley. Plant Cell Physiol. 48 1081–1091. - PubMed
-
- Fuse, T., Sasaki, T., and Yano, M. (2001). Ti-plasmid vectors useful for functional analysis of rice genes. Plant Biotechnol. 18 219–222.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

