Analysis of gene expression and physiological responses in three Mexican maize landraces under drought stress and recovery irrigation
- PMID: 19888455
- PMCID: PMC2766256
- DOI: 10.1371/journal.pone.0007531
Analysis of gene expression and physiological responses in three Mexican maize landraces under drought stress and recovery irrigation
Abstract
Background: Drought is one of the major constraints for plant productivity worldwide. Different mechanisms of drought-tolerance have been reported for several plant species including maize. However, the differences in global gene expression between drought-tolerant and susceptible genotypes and their relationship to physiological adaptations to drought are largely unknown. The study of the differences in global gene expression between tolerant and susceptible genotypes could provide important information to design more efficient breeding programs to produce maize varieties better adapted to water limiting conditions.
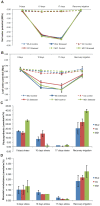
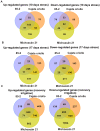
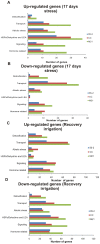
Methodology/principal findings: Changes in physiological responses and gene expression patterns were studied under drought stress and recovery in three Mexican maize landraces which included two drought tolerant (Cajete criollo and Michoacán 21) and one susceptible (85-2) genotypes. Photosynthesis, stomatal conductance, soil and leaf water potentials were monitored throughout the experiment and microarray analysis was carried out on transcripts obtained at 10 and 17 days following application of stress and after recovery irrigation. The two tolerant genotypes show more drastic changes in global gene expression which correlate with different physiological mechanisms of adaptation to drought. Differences in the kinetics and number of up- and down-regulated genes were observed between the tolerant and susceptible maize genotypes, as well as differences between the two tolerant genotypes. Interestingly, the most dramatic differences between the tolerant and susceptible genotypes were observed during recovery irrigation, suggesting that the tolerant genotypes activate mechanisms that allow more efficient recovery after a severe drought.
Conclusions/significance: A correlation between levels of photosynthesis and transcription under stress was observed and differences in the number, type and expression levels of transcription factor families were also identified under drought and recovery between the three maize landraces. Gene expression analysis suggests that the drought tolerant landraces have a greater capacity to rapidly modulate more genes under drought and recovery in comparison to the susceptible landrace. Modulation of a greater number of differentially expressed genes of different TF gene families is an important characteristic of the tolerant genotypes. Finally, important differences were also noted between the tolerant landraces that underlie different mechanisms of achieving tolerance.
Conflict of interest statement
Figures





References
-
- Vinocur B, Altman A. Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Current Opinion in Biotechnology. 2005;16:123–132. - PubMed
-
- Ribaut JM, Ragot M. Marker-assisted selection to improve drought adaptation in maize: the backcross approach, perspectives, limitations, and alternatives. Journal of Experimental Botany. 2007;58:351–360. - PubMed
-
- Aquino P, Carrion F, Calvo R, Flores D. Selected Maize Statistics. Part 4. In: Pingali PL, editor. CIMMYT 1999–2000 World Maize Facts and Trends Meeting World Maize Needs Technological Opportunities and Priorities for the Public Sector. Mexico, D.F.: CIMMYT; 2001. pp. 45–57.
-
- Pingali PL, Pandey S. Meeting World Maize Needs:Technological Opportunities and Priorities for the Public Sector. CIMMYT 1999–2000 World Maize Facts and Trends Meeting World Maize Needs Technological Opportunities and Priorities for the Public Sector 2001
-
- Bruce WB, Edmeades GO, Barker T. Molecular and physiological approaches to maize improvement for drougth tolerance. Journal of Experimental Botany. 2002;53:13–25. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

