Macromolecular micromovements: how RNA polymerase translocates
- PMID: 19889534
- PMCID: PMC3814128
- DOI: 10.1016/j.sbi.2009.10.002
Macromolecular micromovements: how RNA polymerase translocates
Abstract
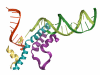
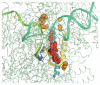
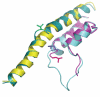
Multi-subunit DNA-dependent RNA polymerases synthesize RNA molecules thousands of nucleotides long. The reiterative reaction of nucleotide condensation occurs at rates of tens of nucleotides per second, invariably linked to the translocation of the enzyme along the DNA template, or threading of the DNA and the nascent RNA molecule through the enzyme. Reiteration of the nucleotide addition/translocation cycle without dissociation from the DNA and RNA requires both isomorphic and metamorphic conformational flexibility of a magnitude substantial enough to accommodate the requisite molecular motions. Here we review some of the more recently acquired insights into the structural flexibility and morphic fluctuations of RNA polymerases and their mechanistic implications.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

