The 5'-untranslated region of the mouse mammary tumor virus mRNA exhibits cap-independent translation initiation
- PMID: 19889724
- PMCID: PMC2811009
- DOI: 10.1093/nar/gkp890
The 5'-untranslated region of the mouse mammary tumor virus mRNA exhibits cap-independent translation initiation
Abstract
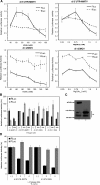
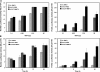
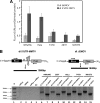
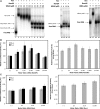
In this study, we demonstrate the identification of an internal ribosome entry site (IRES) within the 5'-untranslated region (5'-UTR) of the mouse mammary tumor virus (MMTV). The 5'-UTR of the full-length mRNA derived from the infectious, complete MMTV genome was cloned into a dual luciferase reporter construct containing an upstream Renilla luciferase gene (RLuc) and a downstream firefly luciferase gene (FLuc). In rabbit reticulocyte lysate, the MMTV 5'-UTR was capable of driving translation of the second cistron. In vitro translational activity from the MMTV 5'-UTR was resistant to the addition of m(7)GpppG cap-analog and cleavage of eIF4G by foot-and-mouth disease virus (FMDV) L-protease. IRES activity was also demonstrated in the Xenopus laevis oocyte by micro-injection of capped and polyadenylated bicistronic RNAs harboring the MMTV-5'-UTR. Finally, transfection assays showed that the MMTV-IRES exhibits cell type-dependent translational activity, suggesting a requirement for as yet unidentified cellular factors for its optimal function.
Figures


References
-
- Callahan R. MMTV-induced mutations in mouse mammary tumors: their potential relevance to human breast cancer. Breast Cancer Res. Treat. 1996;39:33–44. - PubMed
-
- Cohen JC, Varmus HE. Endogenous mammary tumour virus DNA varies among wild mice and segregates during inbreeding. Nature. 1979;278:418–423. - PubMed
-
- Indik S, Gunzburg WH, Salmons B, Rouault F. Mouse mammary tumor virus infects human cells. Cancer Res. 2005;65:6651–6659. - PubMed
-
- Wang Y, Holland JF, Bleiweiss IJ, Melana S, Liu X, Pelisson I, Cantarella A, Stellrecht K, Mani S, Pogo BG. Detection of mammary tumor virus env gene-like sequences in human breast cancer. Cancer Res. 1995;55:5173–5179. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

