Small-molecule activators of a proenzyme
- PMID: 19892984
- PMCID: PMC2886848
- DOI: 10.1126/science.1177585
Small-molecule activators of a proenzyme
Abstract
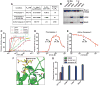
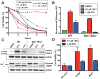
Virtually all of the 560 human proteases are stored as inactive proenyzmes and are strictly regulated. We report the identification and characterization of the first small molecules that directly activate proenzymes, the apoptotic procaspases-3 and -6. It is surprising that these compounds induce autoproteolytic activation by stabilizing a conformation that is both more active and more susceptible to intermolecular proteolysis. These procaspase activators bypass the normal upstream proapoptotic signaling cascades and induce rapid apoptosis in a variety of cell lines. Systematic biochemical and biophysical analyses identified a cluster of mutations in procaspase-3 that resist small-molecule activation both in vitro and in cells. Compounds that induce gain of function are rare, and the activators reported here will enable direct control of the executioner caspases in apoptosis and in cellular differentiation. More generally, these studies presage the discovery of other proenzyme activators to explore fundamental processes of proenzyme activation and their fate-determining roles in biology.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

