Applications of multivariate pattern classification analyses in developmental neuroimaging of healthy and clinical populations
- PMID: 19893761
- PMCID: PMC2773173
- DOI: 10.3389/neuro.09.032.2009
Applications of multivariate pattern classification analyses in developmental neuroimaging of healthy and clinical populations
Abstract
Analyses of functional and structural imaging data typically involve testing hypotheses at each voxel in the brain. However, it is often the case that distributed spatial patterns may be a more appropriate metric for discriminating between conditions or groups. Multivariate pattern analysis has been gaining traction in neuroimaging of adult healthy and clinical populations; studies have shown that information present in neuroimaging data can be used to decode intentions and perceptual states, as well as discriminate between healthy and diseased brains. While few studies to date have applied these methods in pediatric populations, in this review we discuss exciting potential applications for studying both healthy, and aberrant, brain development. We include an overview of methods and discussion of challenges and limitations.
Keywords: MRI; clinical; development; fMRI; multivariate pattern classification.
Figures
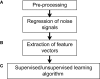
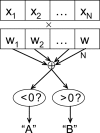
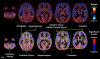
References
-
- Bandettini P. A. (2009). What's New in Neuroimaging Methods? Year in Cognitive Neuroscience 2009. Oxford, Blackwell Publishing, pp. 260–293
-
- Bergstrand S., Bjornsdotter Aberg M., Niiniskorpi T., Wessberg J. (2009). Towards unified analysis of EEG and fMRI: a comparison of classifiers for single-trial pattern recognition. In Proceedings of the Second International Conference on Bio-inspired Systems and Signal Processing, Encarnação P., Veloso A. , eds (Porto, Institute for Systems and Technologies of Information, Control and Communication; ), pp. 273–278
Grants and funding
LinkOut - more resources
Full Text Sources

