DNA methylation pattern changes upon long-term culture and aging of human mesenchymal stromal cells
- PMID: 19895632
- PMCID: PMC2814091
- DOI: 10.1111/j.1474-9726.2009.00535.x
DNA methylation pattern changes upon long-term culture and aging of human mesenchymal stromal cells
Abstract
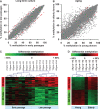
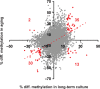
Within 2-3 months of in vitro culture-expansion, mesenchymal stromal cells (MSC) undergo replicative senescence characterized by cell enlargement, loss of differentiation potential and ultimate growth arrest. In this study, we have analyzed DNA methylation changes upon long-term culture of MSC by using the HumanMethylation27 BeadChip microarray assessing 27,578 unique CpG sites. Furthermore, we have compared MSC from young and elderly donors. Overall, methylation patterns were maintained throughout both long-term culture and aging but highly significant differences were observed at specific CpG sites. Many of these differences were observed in homeobox genes and genes involved in cell differentiation. Methylation changes were verified by pyrosequencing after bisulfite conversion and compared to gene expression data. Notably, methylation changes in MSC were overlapping in long-term culture and aging in vivo. This supports the notion that replicative senescence and aging represent developmental processes that are regulated by specific epigenetic modifications.
Figures


References
-
- Banfi A, Muraglia A, Dozin B, Mastrogiacomo M, Cancedda R, Quarto R. Proliferation kinetics and differentiation potential of ex vivo expanded human bone marrow stromal cells: implications for their use in cell therapy. Exp. Hematol. 2000;28:707–715. - PubMed
-
- Baxter MA, Wynn RF, Jowitt SN, Wraith JE, Fairbairn LJ, Bellantuono I. Study of telomere length reveals rapid aging of human marrow stromal cells following in vitro expansion. Stem Cells. 2004;22:675–682. - PubMed
-
- Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435–438. - PubMed
Publication types
MeSH terms
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

