Catalogues of mammalian long noncoding RNAs: modest conservation and incompleteness
- PMID: 19895688
- PMCID: PMC3091318
- DOI: 10.1186/gb-2009-10-11-r124
Catalogues of mammalian long noncoding RNAs: modest conservation and incompleteness
Abstract
Background: Despite increasing interest in the noncoding fraction of transcriptomes, the number, species-conservation and functions, if any, of many non-protein-coding transcripts remain to be discovered. Two extensive long intergenic noncoding RNA (ncRNA) transcript catalogues are now available for mouse: over 3,000 macroRNAs identified by cDNA sequencing, and 1,600 long intergenic noncoding RNA (lincRNA) intervals that are predicted from chromatin-state maps. Previously we showed that macroRNAs tend to be more highly conserved than putatively neutral sequence, although only 5% of bases are predicted as constrained. By contrast, over a thousand lincRNAs were reported as being highly conserved. This apparent difference may account for the surprisingly small fraction (11%) of transcripts that are represented in both catalogues. Here we sought to resolve the reported discrepancy between the evolutionary rates for these two sets.
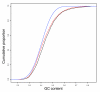
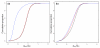
Results: Our analyses reveal lincRNA and macroRNA exon sequences to be subject to the same relatively low degree of sequence constraint. Nonetheless, our observations are consistent with the functionality of a fraction of ncRNA in these sets, with up to a quarter of ncRNA exons having evolved significantly slower than neighboring neutral sequence. The more tissue-specific macroRNAs are enriched in predicted RNA secondary structures and thus may often act in trans, whereas the more highly and broadly expressed lincRNAs appear more likely to act in the cis-regulation of adjacent transcription factor genes.
Conclusions: Taken together, our results indicate that each of the two ncRNA catalogues unevenly and lightly samples the true, much larger, ncRNA repertoire of the mouse.
Figures

References
-
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P, Boyle PJ, Cao H, Carter NP, Clelland GK, Davis S, Day N, Dhami P, Dillon SC, Dorschner MO, Fiegler H. et al.Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. - DOI - PMC - PubMed
-
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. - DOI - PubMed
-
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H. et al.The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. - DOI - PubMed
-
- Wang J, Zhang J, Zheng H, Li J, Liu D, Li H, Samudrala R, Yu J, Wong GK. Mouse transcriptome: neutral evolution of 'non-coding' complementary DNAs. Nature. 2004;431:1. p following 757; discussion following 757. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

