Molecular evolution of multisubunit RNA polymerases: structural analysis
- PMID: 19895816
- PMCID: PMC2813324
- DOI: 10.1016/j.jmb.2009.10.063
Molecular evolution of multisubunit RNA polymerases: structural analysis
Abstract
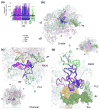
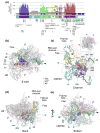
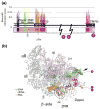
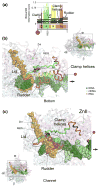
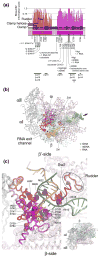
Comprehensive multiple sequence alignments of the multisubunit DNA-dependent RNA polymerase (RNAP) large subunits, including the bacterial beta and beta' subunits and their homologs from archaebacterial RNAPs, eukaryotic RNAPs I-III, nuclear-cytoplasmic large double-stranded DNA virus RNAPs, and plant plastid RNAPs, were created [Lane, W. J. and Darst, S. A. (2009). Molecular evolution of multisubunit RNA polymerases: sequence analysis. In press]. The alignments were used to delineate sequence regions shared among all classes of multisubunit RNAPs, defining common, fundamental RNAP features as well as identifying highly conserved positions. Here, we present a systematic, detailed structural analysis of these shared regions and highly conserved positions in terms of the RNAP structure, as well as the RNAP structure/function relationship, when known.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

