Molecular evolution of multisubunit RNA polymerases: sequence analysis
- PMID: 19895820
- PMCID: PMC2813377
- DOI: 10.1016/j.jmb.2009.10.062
Molecular evolution of multisubunit RNA polymerases: sequence analysis
Abstract
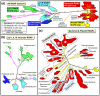
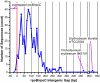
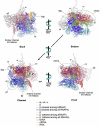
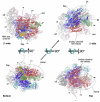
Transcription in all cellular organisms is performed by multisubunit, DNA-dependent RNA polymerases that synthesize RNA from DNA templates. Previous sequence and structural studies have elucidated the importance of shared regions common to all multisubunit RNA polymerases. In addition, RNA polymerases contain multiple lineage-specific domain insertions involved in protein-protein and protein-nucleic acid interactions. We have created comprehensive multiple sequence alignments using all available sequence data for the multisubunit RNA polymerase large subunits, including the bacterial beta and beta' subunits and their homologs from archaebacterial RNA polymerases, the eukaryotic RNA polymerases I, II, and III, the nuclear-cytoplasmic large double-stranded DNA virus RNA polymerases, and plant plastid RNA polymerases. To overcome technical difficulties inherent to the large-subunit sequences, including large sequence length, small and large lineage-specific insertions, split subunits, and fused proteins, we created an automated and customizable sequence retrieval and processing system. In addition, we used our alignments to create a more expansive set of shared sequence regions and bacterial lineage-specific domain insertions. We also analyzed the intergenic gap between the bacterial beta and beta' genes.
Figures





References
-
- Cramer P. Multisubunit RNA polymerases. Curr. Opinion Struct. Biol. 2002;12:89–97. - PubMed
-
- Darst SA. Bacterial RNA polymerase. Curr. Opinion Struct. Biol. 2001;11:155–162. - PubMed
-
- Initiative TAG. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

