Molecular basis for the recognition and cleavages of IGF-II, TGF-alpha, and amylin by human insulin-degrading enzyme
- PMID: 19896952
- PMCID: PMC2813390
- DOI: 10.1016/j.jmb.2009.10.072
Molecular basis for the recognition and cleavages of IGF-II, TGF-alpha, and amylin by human insulin-degrading enzyme
Abstract
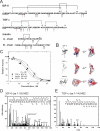
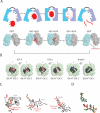
Insulin-degrading enzyme (IDE) is involved in the clearance of many bioactive peptide substrates, including insulin and amyloid-beta, peptides vital to the development of diabetes and Alzheimer's disease, respectively. IDE can also rapidly degrade hormones that are held together by intramolecular disulfide bond(s) without their reduction. Furthermore, IDE exhibits a remarkable ability to preferentially degrade structurally similar peptides such as the selective degradation of insulin-like growth factor (IGF)-II and transforming growth factor-alpha (TGF-alpha) over IGF-I and epidermal growth factor, respectively. Here, we used high-accuracy mass spectrometry to identify the cleavage sites of human IGF-II, TGF-alpha, amylin, reduced amylin, and amyloid-beta by human IDE. We also determined the structures of human IDE-IGF-II and IDE-TGF-alpha at 2.3 A and IDE-amylin at 2.9 A. We found that IDE cleaves its substrates at multiple sites in a biased stochastic manner. Furthermore, the presence of a disulfide bond in amylin allows IDE to cut at an additional site in the middle of the peptide (amino acids 18-19). Our amylin-bound IDE structure offers insight into how the structural constraint from a disulfide bond in amylin can alter IDE cleavage sites. Together with NMR structures of amylin and the IGF and epidermal growth factor families, our work also reveals the structural basis of how the high dipole moment of substrates complements the charge distribution of the IDE catalytic chamber for the substrate selectivity. In addition, we show how the ability of substrates to properly anchor their N-terminus to the exosite of IDE and undergo a conformational switch upon binding to the catalytic chamber of IDE can also contribute to the selective degradation of structurally related growth factors.
Copyright 2009 Elsevier Ltd. All rights reserved.
Figures

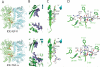

References
-
- Duckworth WC, Bennett RG, Hamel FG. Insulin degradation: progress and potential. Endocr Rev. 1998;19:608–24. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

