The Arabidopsis tandem zinc finger protein AtTZF1 traffics between the nucleus and cytoplasmic foci and binds both DNA and RNA
- PMID: 19897605
- PMCID: PMC2799353
- DOI: 10.1104/pp.109.145656
The Arabidopsis tandem zinc finger protein AtTZF1 traffics between the nucleus and cytoplasmic foci and binds both DNA and RNA
Abstract
Processing bodies (PBs) are specialized cytoplasmic foci where mRNA turnover and translational repression can take place. Stress granules are related cytoplasmic foci. The CCCH tandem zinc finger proteins (TZFs) play pivotal roles in gene expression, cell fate specification, and various developmental processes. Human TZF binds AU-rich elements at the 3' untranslated region and recruits decapping, deadenylation, and exonucleolytic enzymes to PBs for RNA turnover. Recent genetic studies indicate that plant TZFs are involved in gene regulation and hormone-mediated environmental responses. It is unknown if plant TZFs can bind RNA and be localized to PBs or stress granules. The Arabidopsis (Arabidopsis thaliana) AtTZF1/AtCTH/AtC3H23 was identified as a sugar-sensitive gene in a previous microarray study. It is characterized by a TZF motif that is distinct from the human TZF. Higher plants such as Arabidopsis and rice (Oryza sativa) each have a gene family containing this unique TZF motif. Here, we show that AtTZF1 can traffic between the nucleus and cytoplasmic foci. AtTZF1 colocalizes with markers of PBs, and the morphology of these cytoplasmic foci resembles that of mammalian PBs and stress granules. AtTZF1-associated cytoplasmic foci are dynamic and tissue specific. They can be induced by dark and wound stresses and are preferentially present in actively growing tissues and stomatal precursor cells. Since AtTZF1 can bind both DNA and RNA in vitro, it raises the possibility that AtTZF1 might be involved in DNA and/or RNA regulation.
Figures


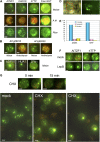



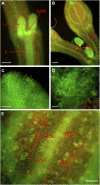
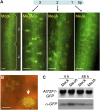


Comment in
-
AtTZF gene family localizes to cytoplasmic foci.Plant Signal Behav. 2010 Feb;5(2):190-2. doi: 10.4161/psb.5.2.10988. Epub 2010 Feb 18. Plant Signal Behav. 2010. PMID: 20173417 Free PMC article.
Similar articles
-
The Arabidopsis thaliana tandem zinc finger 1 (AtTZF1) protein in RNA binding and decay.Plant J. 2014 May;78(3):452-67. doi: 10.1111/tpj.12485. Epub 2014 Apr 15. Plant J. 2014. PMID: 24635033 Free PMC article.
-
The Arabidopsis tandem zinc finger protein AtTZF1 affects ABA- and GA-mediated growth, stress and gene expression responses.Plant J. 2011 Jan;65(2):253-68. doi: 10.1111/j.1365-313X.2010.04419.x. Epub 2010 Dec 8. Plant J. 2011. PMID: 21223390
-
Can AtTZF1 act as a transcriptional activator or repressor in plants?Plant Signal Behav. 2011 May;6(5):719-22. doi: 10.4161/psb.6.5.15104. Epub 2011 May 1. Plant Signal Behav. 2011. PMID: 21455027 Free PMC article.
-
Arginine-rich motif-tandem CCCH zinc finger proteins in plant stress responses and post-transcriptional regulation of gene expression.Plant Sci. 2016 Nov;252:118-124. doi: 10.1016/j.plantsci.2016.06.014. Epub 2016 Jul 21. Plant Sci. 2016. PMID: 27717446 Review.
-
Tandem CCCH zinc finger proteins in plant growth, development and stress response.Plant Cell Physiol. 2014 Aug;55(8):1367-75. doi: 10.1093/pcp/pcu074. Epub 2014 May 21. Plant Cell Physiol. 2014. PMID: 24850834 Review.
Cited by
-
Conservation of AtTZF1, AtTZF2, and AtTZF3 homolog gene regulation by salt stress in evolutionarily distant plant species.Front Plant Sci. 2015 Jun 16;6:394. doi: 10.3389/fpls.2015.00394. eCollection 2015. Front Plant Sci. 2015. PMID: 26136754 Free PMC article.
-
The Plasmodium falciparum CCCH Zinc Finger Protein ZNF4 Plays an Important Role in Gametocyte Exflagellation through the Regulation of Male Enriched Transcripts.Cells. 2022 May 17;11(10):1666. doi: 10.3390/cells11101666. Cells. 2022. PMID: 35626703 Free PMC article.
-
STRONG STAYGREEN inhibits DNA binding of PvNAP transcription factors during leaf senescence in switchgrass.Plant Physiol. 2022 Oct 27;190(3):2045-2058. doi: 10.1093/plphys/kiac397. Plant Physiol. 2022. PMID: 36005925 Free PMC article.
-
Connecting the dots of RNA-directed DNA methylation in Arabidopsis thaliana.Chromosome Res. 2014 Jun;22(2):225-40. doi: 10.1007/s10577-014-9425-9. Chromosome Res. 2014. PMID: 24846724 Review.
-
Composition and function of P bodies in Arabidopsis thaliana.Front Plant Sci. 2014 May 14;5:201. doi: 10.3389/fpls.2014.00201. eCollection 2014. Front Plant Sci. 2014. PMID: 24860588 Free PMC article. Review.
References
-
- Anderson P, Kedersha N (2008) Stress granules: the tao of RNA triage. Trends Biochem Sci 33 141–150 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

