P1 plasmid segregation: accurate redistribution by dynamic plasmid pairing and separation
- PMID: 19897644
- PMCID: PMC2820840
- DOI: 10.1128/JB.01245-09
P1 plasmid segregation: accurate redistribution by dynamic plasmid pairing and separation
Abstract
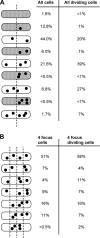
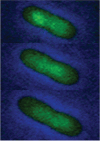
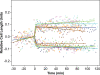
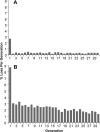
Low-copy-number plasmids, such as P1 and F, encode a type Ia partition system (P1par or Fsop) for active segregation of copies to daughter cells. Typical descriptions show a single central plasmid focus dividing and the products moving to the cell quarter regions, ensuring segregation. However, using improved optical and analytical tools and large cell populations, we show that P1 plasmid foci are very broadly distributed. Moreover, under most growth conditions, more than two foci are frequently present. Each focus contains either one or two plasmid copies. Replication and focus splitting occur at almost any position in the cell. The products then move rapidly apart for approximately 40% of the cell length. They then tend to maintain their relative positions. The segregating foci often pass close to or come to rest close to other foci in the cell. Foci frequently appear to fuse during these encounters. Such events occur several times in each cell and cell generation on average. We argue that foci pair with their neighbors and then actively separate again. The net result is an approximately even distribution of foci along the long cell axis on average. We show mathematically that trans-pairing and active separation could greatly increase the accuracy of segregation and would produce the distributions of foci that we observe. Plasmid pairing and separation may constitute a novel fine-tuning mechanism that takes the basic pattern created when plasmids separate after replication and converts it to a roughly even pattern that greatly improves the fidelity of plasmid segregation.
Figures





Comment in
-
Plasmid segregation: birds of a feather try not to flock together.J Bacteriol. 2010 Mar;192(5):1171-4. doi: 10.1128/JB.01551-09. Epub 2009 Dec 18. J Bacteriol. 2010. PMID: 20023023 Free PMC article. No abstract available.
References
-
- Adachi, S., K. Hori, and S. Hiraga. 2006. Subcellular positioning of F plasmid mediated by dynamic localization of SopA and SopB. J. Mol. Biol. 356:850-863. - PubMed
-
- Castaing, J. P., J. Y. Bouet, and D. Lane. 2008. F plasmid partition depends on interaction of SopA with non-specific DNA. Mol. Microbiol. 70:1000-1011. - PubMed
-
- Davis, M. A., K. A. Martin, and S. J. Austin. 1992. Biochemical activities of the ParA partition protein of the P1 plasmid. Mol. Microbiol. 6:1141-1147. - PubMed
-
- Davis, M. A., L. Radnedge, K. A. Martin, F. Hayes, B. Youngren, and S. J. Austin. 1996. The P1 ParA protein and its ATPase activity play a direct role in the segregation of plasmid copies to daughter cells. Mol. Microbiol. 21:1029-1036. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

