Functional copies of the Mst77F gene on the Y chromosome of Drosophila melanogaster
- PMID: 19897751
- PMCID: PMC2815925
- DOI: 10.1534/genetics.109.107516
Functional copies of the Mst77F gene on the Y chromosome of Drosophila melanogaster
Abstract
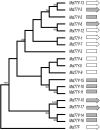
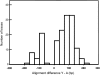
The Y chromosome of Drosophila melanogaster has <20 protein-coding genes. These genes originated from the duplication of autosomal genes and have male-related functions. In 1993, Russell and Kaiser found three Y-linked pseudogenes of the Mst77F gene, which is a testis-expressed autosomal gene that is essential for male fertility. We did a thorough search using experimental and computational methods and found 18 Y-linked copies of this gene (named Mst77Y-1-Mst77Y-18). Ten Mst77Y genes encode defective proteins and the other eight are potentially functional. These eight genes produce approximately 20% of the functional Mst77F-like mRNA, and molecular evolutionary analysis shows that they evolved under purifying selection. Hence several Mst77Y genes have all the features of functional genes. Mst77Y genes are present only in D. melanogaster, and phylogenetic analysis confirmed that the duplication is a recent event. The identification of functional Mst77Y genes reinforces the previous finding that gene gains play a prominent role in the evolution of the Drosophila Y chromosome.
Figures


References
-
- Abad, J. P., B. de Pablos, M. Agudo, I. Molina, G. Giovinazzo et al., 2004. Genomic and cytological analysis of the Y chromosome of Drosophila melanogaster: telomere-derived sequences at internal regions. Chromosoma 113 295–304. - PubMed
-
- Adams, M. D., S. E. Celniker, R. A. Holt, C. A. Evans, J. D. Gocayne et al., 2000. The genome sequence of Drosophila melanogaster. Science 287 2185–2195. - PubMed
-
- Bailey, J. A., Z. Gu, R. A. Clark, K. Reinert, R. V. Samonte et al., 2002. Recent segmental duplications in the human genome. Science 297 1003–1007. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

