Dancing genomes: fungal nuclear positioning
- PMID: 19898490
- PMCID: PMC2794368
- DOI: 10.1038/nrmicro2249
Dancing genomes: fungal nuclear positioning
Abstract
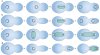
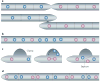
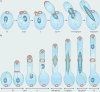
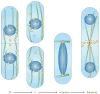
The many different mechanisms that fungi use to transmit and share genetic material are mediated by a broad range of chromosome and nuclear dynamics. The mechanics underlying nuclear migration are well integrated into detailed models, in which the forces supplied by plus- and minus-end-directed microtubule motors position and move the nucleus in a cell. Although we know much about how cells move nuclei, we know much less about why the cell invests in so many different nuclear 'dances'. Here, we briefly survey the available models for the mechanics of nuclear migration in fungi and then focus on examples of how fungal cells use these nuclear dances - the movement of intact nuclei in and between cells - to control the integrity, ploidy and assortment of specific genomes or individual chromosomes.
Figures

References
-
- Petersen J, Heitz MJ, Hagan IM. Conjugation in S. pombe: identification of a microtubule-organising centre, a requirement for microtubules and a role for Mad2. Curr Biol. 1998;8:963–966. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

