Chromosome 7 and 19 trisomy in cultured human neural progenitor cells
- PMID: 19898616
- PMCID: PMC2765070
- DOI: 10.1371/journal.pone.0007630
Chromosome 7 and 19 trisomy in cultured human neural progenitor cells
Abstract
Background: Stem cell expansion and differentiation is the foundation of emerging cell therapy technologies. The potential applications of human neural progenitor cells (hNPCs) are wide ranging, but a normal cytogenetic profile is important to avoid the risk of tumor formation in clinical trials. FDA approved clinical trials are being planned and conducted for hNPC transplantation into the brain or spinal cord for various neurodegenerative disorders. Although human embryonic stem cells (hESCs) are known to show recurrent chromosomal abnormalities involving 12 and 17, no studies have revealed chromosomal abnormalities in cultured hNPCs. Therefore, we investigated frequently occurring chromosomal abnormalities in 21 independent fetal-derived hNPC lines and the possible mechanisms triggering such aberrations.
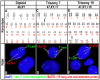
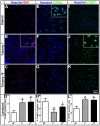
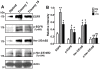
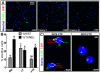
Methods and findings: While most hNPC lines were karyotypically normal, G-band karyotyping and fluorescent in situ hybridization (FISH) analyses revealed the emergence of trisomy 7 (hNPC(+7)) and trisomy 19 (hNPC(+19)), in 24% and 5% of the lines, respectively. Once detected, subsequent passaging revealed emerging dominance of trisomy hNPCs. DNA microarray and immunoblotting analyses demonstrate epidermal growth factor receptor (EGFR) overexpression in hNPC(+7) and hNPC(+19) cells. We observed greater levels of telomerase (hTERT), increased proliferation (Ki67), survival (TUNEL), and neurogenesis (beta(III)-tubulin) in hNPC(+7) and hNPC(+19), using respective immunocytochemical markers. However, the trisomy lines underwent replicative senescence after 50-60 population doublings and never showed neoplastic changes. Although hNPC(+7) and hNPC(+19) survived better after xenotransplantation into the rat striatum, they did not form malignant tumors. Finally, EGF deprivation triggered a selection of trisomy 7 cells in a diploid hNPC line.
Conclusions: We report that hNPCs are susceptible to accumulation of chromosome 7 and 19 trisomy in long-term cell culture. These results suggest that micro-environmental cues are powerful factors in the selection of specific hNPC aneuploidies, with trisomy of chromosome 7 being the most common. Given that a number of stem cell based clinical trials are being conducted or planned in USA and a recent report in PLoS Medicine showing the dangers of grafting an inordinate number of cells, these data substantiate the need for careful cytogenetic evaluation of hNPCs (fetal or hESC-derived) before their use in clinical or basic science applications.
Conflict of interest statement
Figures
References
-
- Hoffman LM, Carpenter MK. Characterization and culture of human embryonic stem cells. Nat Biotechnol. 2005;23(6):699–708. - PubMed
-
- Baker DE, Harrison NJ, Maltby E, Smith K, Moore HD, et al. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat Biotechnol. 2007;25(2):207–215. - PubMed
-
- Draper JS, Smith K, Gokhale P, Moore HD, Maltby E, et al. Recurrent gain of chromosomes 17q and 12 in cultured human embryonic stem cells. Nat Biotechnol. 2004;22(1):53–54. - PubMed
-
- Spits C, Mateizel I, Geens M, Mertzanidou A, Staessen C, et al. Recurrent chromosomal abnormalities in human embryonic stem cells. Nat Biotechnol. 2008;26(12):1361–1363. - PubMed
-
- Meisner LF, Johnson JA. Protocols for cytogenetic studies of human embryonic stem cells. Methods. 2008;45(2):133–141. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

