DNA barcode analysis: a comparison of phylogenetic and statistical classification methods
- PMID: 19900297
- PMCID: PMC2775147
- DOI: 10.1186/1471-2105-10-S14-S10
DNA barcode analysis: a comparison of phylogenetic and statistical classification methods
Abstract
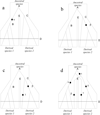
Background: DNA barcoding aims to assign individuals to given species according to their sequence at a small locus, generally part of the CO1 mitochondrial gene. Amongst other issues, this raises the question of how to deal with within-species genetic variability and potential transpecific polymorphism. In this context, we examine several assignation methods belonging to two main categories: (i) phylogenetic methods (neighbour-joining and PhyML) that attempt to account for the genealogical framework of DNA evolution and (ii) supervised classification methods (k-nearest neighbour, CART, random forest and kernel methods). These methods range from basic to elaborate. We investigated the ability of each method to correctly classify query sequences drawn from samples of related species using both simulated and real data. Simulated data sets were generated using coalescent simulations in which we varied the genealogical history, mutation parameter, sample size and number of species.
Results: No method was found to be the best in all cases. The simplest method of all, "one nearest neighbour", was found to be the most reliable with respect to changes in the parameters of the data sets. The parameter most influencing the performance of the various methods was molecular diversity of the data. Addition of genetically independent loci--nuclear genes--improved the predictive performance of most methods.
Conclusion: The study implies that taxonomists can influence the quality of their analyses either by choosing a method best-adapted to the configuration of their sample, or, given a certain method, increasing the sample size or altering the amount of molecular diversity. This can be achieved either by sequencing more mtDNA or by sequencing additional nuclear genes. In the latter case, they may also have to modify their data analysis method.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

