Members of a novel protein family containing microneme adhesive repeat domains act as sialic acid-binding lectins during host cell invasion by apicomplexan parasites
- PMID: 19901027
- PMCID: PMC2804363
- DOI: 10.1074/jbc.M109.060988
Members of a novel protein family containing microneme adhesive repeat domains act as sialic acid-binding lectins during host cell invasion by apicomplexan parasites
Abstract
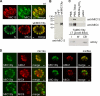
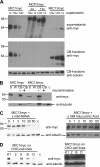
Numerous intracellular pathogens exploit cell surface glycoconjugates for host cell recognition and entry. Unlike bacteria and viruses, Toxoplasma gondii and other parasites of the phylum Apicomplexa actively invade host cells, and this process critically depends on adhesins (microneme proteins) released onto the parasite surface from intracellular organelles called micronemes (MIC). The microneme adhesive repeat (MAR) domain of T. gondii MIC1 (TgMIC1) recognizes sialic acid (Sia), a key determinant on the host cell surface for invasion by this pathogen. By complementation and invasion assays, we demonstrate that TgMIC1 is one important player in Sia-dependent invasion and that another novel Sia-binding lectin, designated TgMIC13, is also involved. Using BLAST searches, we identify a family of MAR-containing proteins in enteroparasitic coccidians, a subclass of apicomplexans, including T. gondii, suggesting that all these parasites exploit sialylated glycoconjugates on host cells as determinants for enteric invasion. Furthermore, this protein family might provide a basis for the broad host cell range observed for coccidians that form tissue cysts during chronic infection. Carbohydrate microarray analyses, corroborated by structural considerations, show that TgMIC13, TgMIC1, and its homologue Neospora caninum MIC1 (NcMIC1) share a preference for alpha2-3- over alpha2-6-linked sialyl-N-acetyllactosamine sequences. However, the three lectins also display differences in binding preferences. Intense binding of TgMIC13 to alpha2-9-linked disialyl sequence reported on embryonal cells and relatively strong binding to 4-O-acetylated-Sia found on gut epithelium and binding of NcMIC1 to 6'sulfo-sialyl Lewis(x) might have implications for tissue tropism.
Figures




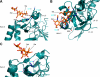
References
-
- Takabatake N., Okamura M., Yokoyama N., Ikehara Y., Akimitsu N., Arimitsu N., Hamamoto H., Sekimizu K., Suzuki H., Igarashi I. (2007) Vet. Parasitol. 148, 93–101 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

