Epidemic multiple drug resistant Salmonella Typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype
- PMID: 19901036
- PMCID: PMC2792184
- DOI: 10.1101/gr.091017.109
Epidemic multiple drug resistant Salmonella Typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype
Abstract
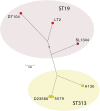
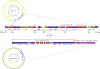
Whereas most nontyphoidal Salmonella (NTS) are associated with gastroenteritis, there has been a dramatic increase in reports of NTS-associated invasive disease in sub-Saharan Africa. Salmonella enterica serovar Typhimurium isolates are responsible for a significant proportion of the reported invasive NTS in this region. Multilocus sequence analysis of invasive S. Typhimurium from Malawi and Kenya identified a dominant type, designated ST313, which currently is rarely reported outside of Africa. Whole-genome sequencing of a multiple drug resistant (MDR) ST313 NTS isolate, D23580, identified a distinct prophage repertoire and a composite genetic element encoding MDR genes located on a virulence-associated plasmid. Further, there was evidence of genome degradation, including pseudogene formation and chromosomal deletions, when compared with other S. Typhimurium genome sequences. Some of this genome degradation involved genes previously implicated in virulence of S. Typhimurium or genes for which the orthologs in S. Typhi are either pseudogenes or are absent. Genome analysis of other epidemic ST313 isolates from Malawi and Kenya provided evidence for microevolution and clonal replacement in the field.
Figures


References
-
- Berkley JA, Lowe BS, Mwangi I, Williams T, Bauni E, Mwarumba S, Ngetsa C, Slack MP, Njenga S, Hart CA, et al. Bacteremia among children admitted to a rural hospital in Kenya. N Engl J Med. 2005;352:39–47. - PubMed
-
- Brent AJ, Oundo JO, Mwangi I, Ochola L, Lowe B, Berkley JA. Salmonella bacteremia in Kenyan children. Pediatr Infect Dis J. 2006;25:230–236. - PubMed
-
- Cooke FJ, Wain J, Fookes M, Ivens A, Thomson N, Brown DJ, Threlfall EJ, Gunn G, Foster G, Dougan G. Prophage sequences defining hot spots of genome variation in Salmonella enterica serovar Typhimurium can be used to discriminate between field isolates. J Clin Microbiol. 2007;45:2590–2598. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
