Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA
- PMID: 19901979
- PMCID: PMC2766070
- DOI: 10.1371/journal.pbio.1000238
Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA
Abstract
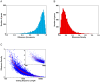
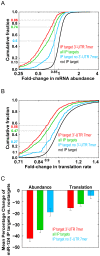
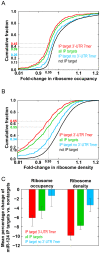
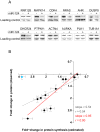
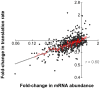
MicroRNAs (miRNAs) regulate gene expression posttranscriptionally by interfering with a target mRNA's translation, stability, or both. We sought to dissect the respective contributions of translational inhibition and mRNA decay to microRNA regulation. We identified direct targets of a specific miRNA, miR-124, by virtue of their association with Argonaute proteins, core components of miRNA effector complexes, in response to miR-124 transfection in human tissue culture cells. In parallel, we assessed mRNA levels and obtained translation profiles using a novel global approach to analyze polysomes separated on sucrose gradients. Analysis of translation profiles for approximately 8,000 genes in these proliferative human cells revealed that basic features of translation are similar to those previously observed in rapidly growing Saccharomyces cerevisiae. For approximately 600 mRNAs specifically recruited to Argonaute proteins by miR-124, we found reductions in both the mRNA abundance and inferred translation rate spanning a large dynamic range. The changes in mRNA levels of these miR-124 targets were larger than the changes in translation, with average decreases of 35% and 12%, respectively. Further, there was no identifiable subgroup of mRNA targets for which the translational response was dominant. Both ribosome occupancy (the fraction of a given gene's transcripts associated with ribosomes) and ribosome density (the average number of ribosomes bound per unit length of coding sequence) were selectively reduced for hundreds of miR-124 targets by the presence of miR-124. Changes in protein abundance inferred from the observed changes in mRNA abundance and translation profiles closely matched changes directly determined by Western analysis for 11 of 12 proteins, suggesting that our assays captured most of miR-124-mediated regulation. These results suggest that miRNAs inhibit translation initiation or stimulate ribosome drop-off preferentially near the start site and are not consistent with inhibition of polypeptide elongation, or nascent polypeptide degradation contributing significantly to miRNA-mediated regulation in proliferating HEK293T cells. The observation of concordant changes in mRNA abundance and translational rate for hundreds of miR-124 targets is consistent with a functional link between these two regulatory outcomes of miRNA targeting, and the well-documented interrelationship between translation and mRNA decay.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Lee R. C, Feinbaum R. L, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. - PubMed
-
- Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Filipowicz W, Bhattacharyya S. N, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. - PubMed
-
- Lewis B. P, Burge C. B, Bartel D. P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

