Visualization of subcellular NAD pools and intra-organellar protein localization by poly-ADP-ribose formation
- PMID: 19902144
- PMCID: PMC11115696
- DOI: 10.1007/s00018-009-0190-4
Visualization of subcellular NAD pools and intra-organellar protein localization by poly-ADP-ribose formation
Abstract
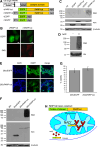
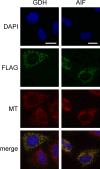
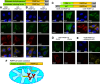
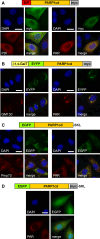
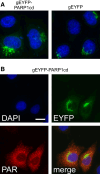
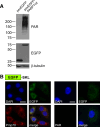
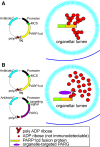
Poly-ADP-ribose polymerases (PARPs) use NAD(+) as substrate to generate polymers of ADP-ribose. We targeted the catalytic domain of human PARP1 as molecular NAD(+) detector into cellular organelles. Immunochemical detection of polymers demonstrated distinct subcellular NAD(+) pools in mitochondria, peroxisomes and, surprisingly, in the endoplasmic reticulum and the Golgi complex. Polymers did not accumulate within the mitochondrial intermembrane space or the cytosol. We demonstrate the suitability of this compartment-specific NAD(+) and poly-ADP-ribose turnover to establish intra-organellar protein localization. For overexpressed proteins, genetically endowed with PARP activity, detection of polymers indicates segregation from the cytosol and consequently intra-organellar residence. In mitochondria, polymer build-up reveals matrix localization of the PARP fusion protein. Compared to presently used fusion tags for subcellular protein localization, these are substantial improvements in resolution. We thus established a novel molecular tool applicable for studies of subcellular NAD metabolism and protein localization.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

