Activating mutations in TOR are in similar structures as oncogenic mutations in PI3KCalpha
- PMID: 19902965
- PMCID: PMC2796128
- DOI: 10.1021/cb900193e
Activating mutations in TOR are in similar structures as oncogenic mutations in PI3KCalpha
Abstract
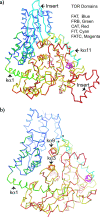
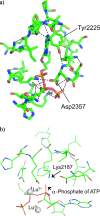
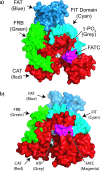
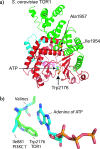
TOR (Target of Rapamycin) is a highly conserved Ser/Thr kinase and a central controller of cell growth. Using the crystal structure of the related lipid kinase PI3KCgamma, we built a model of the catalytic region of TOR, from the FAT domain to near the end of the FATC domain. The model reveals that activating mutations in TOR, identified in yeast in a genetic selection for Rheb-independence, correspond to hotspots for oncogenic mutations in PI3KCalpha. The activating mutations are in the catalytic domain (helices kalpha3, kalpha9, kalpha11) and the helical domain of TOR. Docking studies with small molecule inhibitors (PP242, NVP-BEZ235, and Ku-0063794) show that drugs currently in development utilize a novel pharmacophore space to achieve specificity. Thus, our model provides insight on the regulation of TOR and may be useful in the design of new anticancer drugs.
Figures

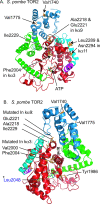
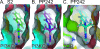
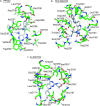
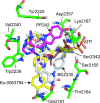

References
-
- Brachmann S.; Fritsch C.; Maira S. M.; Garcia-Echeverria C. (2009) PI3K and mTOR inhibitors: a new generation of targeted anticancer agents. Curr. Opin. Cell Biol. 21, 194–198. - PubMed
-
- Wullschleger S.; Loewith R.; Hall M. N. (2006) TOR signaling in growth and metabolism. Cell 124, 471–484. - PubMed
-
- Guertin D. A.; Sabatini D. M. (2007) Defining the role of mTOR in cancer. Cancer Cell 12, 9–22. - PubMed
-
- Huang C. H.; Mandelker D.; Schmidt-Kittler O.; Samuels Y.; Velculescu V. E.; Kinzler K. W.; Vogelstein B.; Gabelli S. B.; Amzel L. M. (2007) The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations. Science (New York, N.Y.) 318, 1744–1748. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

