Genomic events underlying the changes in adamantane resistance among influenza A(H3N2) viruses during 2006-2008
- PMID: 19903212
- PMCID: PMC4984730
- DOI: 10.1111/j.1750-2659.2009.00103.x
Genomic events underlying the changes in adamantane resistance among influenza A(H3N2) viruses during 2006-2008
Abstract
Background: Adamantanes resistance in H3N2 viruses has been increasing since 2000, and in 2005-2006 reached nearly 100% in most countries, with the circulation of the N-lineage. In 2006-2007, however, a significant decrease in resistance was observed in many regions.
Objectives: To explore potential links between adamantane resistance and the A(H3N2) viruses that circulated between 2006 and 2008.
Methods: A total of 1451 Influenza A (H3N2) viruses collected globally in 2001-2008 were screened for the presence of adamantane resistance markers. A subset of 100 viruses representing the broad genetic and geographic spectrum of these viruses was selected for complete genome sequencing and phylogenetic analyses.
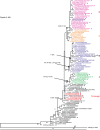
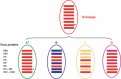
Results: Full genome sequence analysis of 2006-2007 viruses revealed co-circulation of four distinct genotypes, designated A-D. Phylogenetic analyses demonstrated reassortment between viruses from the N-lineage and other viruses that had circulated in prior seasons, including those bearing an adamantane sensitive marker. Genotype D viruses became dominant in late 2006-2007 and continued to be the main H3N2 genotype in 2007-2008. Viruses of this genotype retained all N-lineage genome segments except PB2 and NP, which were acquired through reassortment.
Conclusions: The decrease in adamantane resistance at that time was due to transient co-circulation of genotypes that emerged through reassortment. Our findings emphasize the importance of complete genome sequencing in understanding the complex nature of the relationship between influenza virus evolution and antiviral resistance. The recent emergence of the pandemic multi-reassortant H1N1 virus underscores the importance of whole genome sequence monitoring for rapid detection of such unusual and novel strains.
Figures









References
-
- Hay AJ, Zambon MC, Wolstenholme AJ, Skehel JJ, Smith MH. Molecular basis of resistance of influenza A viruses to amantadine. J Antimicrob Chemother 1986; 18(Suppl B):19–29. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

