Performance of optimized McRAPD in identification of 9 yeast species frequently isolated from patient samples: potential for automation
- PMID: 19903328
- PMCID: PMC2779194
- DOI: 10.1186/1471-2180-9-234
Performance of optimized McRAPD in identification of 9 yeast species frequently isolated from patient samples: potential for automation
Abstract
Background: Rapid, easy, economical and accurate species identification of yeasts isolated from clinical samples remains an important challenge for routine microbiological laboratories, because susceptibility to antifungal agents, probability to develop resistance and ability to cause disease vary in different species. To overcome the drawbacks of the currently available techniques we have recently proposed an innovative approach to yeast species identification based on RAPD genotyping and termed McRAPD (Melting curve of RAPD). Here we have evaluated its performance on a broader spectrum of clinically relevant yeast species and also examined the potential of automated and semi-automated interpretation of McRAPD data for yeast species identification.
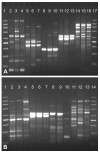
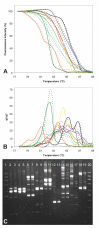
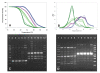
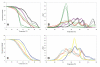
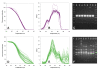
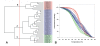
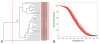
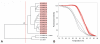
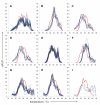
Results: A simple fully automated algorithm based on normalized melting data identified 80% of the isolates correctly. When this algorithm was supplemented by semi-automated matching of decisive peaks in first derivative plots, 87% of the isolates were identified correctly. However, a computer-aided visual matching of derivative plots showed the best performance with average 98.3% of the accurately identified isolates, almost matching the 99.4% performance of traditional RAPD fingerprinting.
Conclusion: Since McRAPD technique omits gel electrophoresis and can be performed in a rapid, economical and convenient way, we believe that it can find its place in routine identification of medically important yeasts in advanced diagnostic laboratories that are able to adopt this technique. It can also serve as a broad-range high-throughput technique for epidemiological surveillance.
Figures






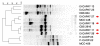
References
-
- Trtkova J, Raclavsky V. Molecular-genetic approaches to identification and typing of pathogenic Candida yeasts. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2006;150:51–61. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

