Comparative analysis of Panicum streak virus and Maize streak virus diversity, recombination patterns and phylogeography
- PMID: 19903330
- PMCID: PMC2777162
- DOI: 10.1186/1743-422X-6-194
Comparative analysis of Panicum streak virus and Maize streak virus diversity, recombination patterns and phylogeography
Abstract
Background: Panicum streak virus (PanSV; Family Geminiviridae; Genus Mastrevirus) is a close relative of Maize streak virus (MSV), the most serious viral threat to maize production in Africa. PanSV and MSV have the same leafhopper vector species, largely overlapping natural host ranges and similar geographical distributions across Africa and its associated Indian Ocean Islands. Unlike MSV, however, PanSV has no known economic relevance.
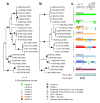
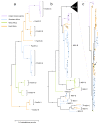
Results: Here we report on 16 new PanSV full genome sequences sampled throughout Africa and use these together with others in public databases to reveal that PanSV and MSV populations in general share very similar patterns of genetic exchange and geographically structured diversity. A potentially important difference between the species, however, is that the movement of MSV strains throughout Africa is apparently less constrained than that of PanSV strains. Interestingly the MSV-A strain which causes maize streak disease is apparently the most mobile of all the PanSV and MSV strains investigated.
Conclusion: We therefore hypothesize that the generally increased mobility of MSV relative to other closely related species such as PanSV, may have been an important evolutionary step in the eventual emergence of MSV-A as a serious agricultural pathogen.The GenBank accession numbers for the sequences reported in this paper are GQ415386-GQ415401.
Figures
References
-
- Pinner MS, Markham PG, Markham RH, Dekker EL. Characterization of maize streak virus:description of strains; symptoms. Plant Path. 1988;6:74–87. doi: 10.1111/j.1365-3059.1988.tb02198.x. - DOI
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

