Genome Alteration Print (GAP): a tool to visualize and mine complex cancer genomic profiles obtained by SNP arrays
- PMID: 19903341
- PMCID: PMC2810663
- DOI: 10.1186/gb-2009-10-11-r128
Genome Alteration Print (GAP): a tool to visualize and mine complex cancer genomic profiles obtained by SNP arrays
Abstract
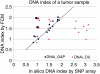
We describe a method for automatic detection of absolute segmental copy numbers and genotype status in complex cancer genome profiles measured with single-nucleotide polymorphism (SNP) arrays. The method is based on pattern recognition of segmented and smoothed copy number and allelic imbalance profiles. Assignments were verified by DNA indexes of primary tumors and karyotypes of cell lines. The method performs well even for poor-quality data, low tumor content, and highly rearranged tumor genomes.
Figures




References
-
- Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, Lapuk A, Neve RM, Qian Z, Ryder T, Chen F, Feiler H, Tokuyasu T, Kingsley C, Dairkee S, Meng Z, Chew K, Pinkel D, Jain A, Ljung BM, Esserman L, Albertson DG, Waldman FM, Gray JW. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

