Evolving views of DNA replication (in)fidelity
- PMID: 19903750
- PMCID: PMC3628614
- DOI: 10.1101/sqb.2009.74.027
Evolving views of DNA replication (in)fidelity
Abstract
"It has not escaped our notice that the specific pairing we have postulated immediately suggests a possible copying mechanism for the genetic material" (Watson and Crick 1953). In the years since this remarkable understatement, we have come to realize the enormous complexity of the cellular machinery devoted to replicating DNA with the accuracy needed to maintain genetic information over many generations, balanced by the emergence of mutations on which selection can act. This complexity is partly based on the need to remove or tolerate cytotoxic and mutagenic lesions in DNA generated by environmental stress. Considered here is the fidelity with which undamaged and damaged DNA is replicated by the many DNA polymerases now known to exist. Some of these seriously violate Watson-Crick base-pairing rules such that, depending on the polymerase, the composition and location of the error, and the ability to correct errors (or not), DNA synthesis error rates can vary by more than a millionfold. This offers the potential to modulate rates of point mutations over a wide range, with consequences that can be either deleterious or beneficial.
Figures
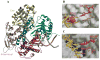

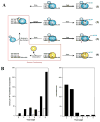
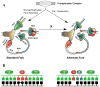
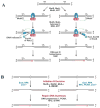
References
-
- Beard WA, Wilson SH. Structural insights into the origins of DNA polymerase fidelity. Structure 2003 - PubMed
-
- Bebenek K, Kunkel TA. Streisinger revisited: DNA synthesis errors mediated by substrate misalignments. Cold Spring Harb Symp Quant Biol. 2000;65:81–91. - PubMed
-
- Bebenek K, Kunkel TA, editors. Functions of DNA Polymerases in DNA Repair and Replication. 2004. - PubMed
-
- Bebenek K, Tissier A, Frank EG, McDonald JP, Prasad R, Wilson SH, Woodgate R, Kunkel TA. 5′-Deoxyribose phosphate lyase activity of human DNA polymerase iota in vitro. Science (New York, NY. 2001;291:2156–2159. - PubMed
-
- Bessman MJ, Kornberg A, Lehman IR, Simms ES. Enzymic synthesis of deoxyribonucleic acid. Biochim Biophys Acta. 1956;21:197–198. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
