A conserved transcriptional regulator is required for RNA-directed DNA methylation and plant development
- PMID: 19903758
- PMCID: PMC2788325
- DOI: 10.1101/gad.1851809
A conserved transcriptional regulator is required for RNA-directed DNA methylation and plant development
Abstract
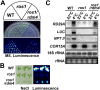
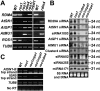
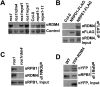
RNA-directed DNA methylation (RdDM) is a conserved mechanism for epigenetic silencing of transposons and other repetitive elements. We report that the rdm4 (RNA-directed DNA Methylation4) mutation not only impairs RdDM, but also causes pleiotropic developmental defects in Arabidopsis. Both RNA polymerase II (Pol II)- and Pol V-dependent transcripts are affected in the rdm4 mutant. RDM4 encodes a novel protein that is conserved from yeast to humans and interacts with Pol II and Pol V in plants. Our results suggest that RDM4 functions in epigenetic regulation and plant development by serving as a transcriptional regulator for RNA Pol V and Pol II, respectively.
Figures


References
-
- Chan SW, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet. 2005;6:351–360. - PubMed
-
- Cramer P. Multisubunit RNA polymerases. Curr Opin Struct Biol. 2002;12:89–97. - PubMed
-
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. - PubMed
-
- Gong Z, Morales-Ruiz T, Ariza RR, Roldan-Arjona T, David L, Zhu JK. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–814. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
