Rapid automated cell quantification on HIV microfluidic devices
- PMID: 19904402
- PMCID: PMC3839566
- DOI: 10.1039/b911882a
Rapid automated cell quantification on HIV microfluidic devices
Abstract
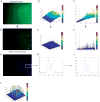
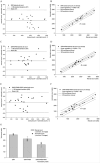
Lab-chip device analysis often requires high throughput quantification of fluorescent cell images, obtained under different conditions of fluorescent intensity, illumination, focal depth, and optical magnification. Many laboratories still use manual counting--a tedious, expensive process prone to inter-observer variability. The manual counting process can be automated for fast and precise data gathering and reduced manual bias. We present a method to segment and count cells in microfluidic chips that are labeled with a single stain, or multiple stains, using image analysis techniques in Matlab and discuss its advantages over manual counting. Microfluidic based cell capturing devices for HIV monitoring were used to validate our method. Captured CD4(+) CD3(+) T lymphocytes were stained with DAPI, AF488-anti CD4, and AF647-anti CD3 for cell identification. Altogether 4788 (76 x 3 x 21) gray color images were obtained from devices using discarded 10 HIV infected patient whole blood samples (21 devices). We observed that the automatic method performs similarly to manual counting for a small number of cells. However, automated counting is more accurate and more than 100 times faster than manual counting for multiple-color stained cells, especially when large numbers of cells need to be quantified (>500 cells). The algorithm is fully automatic for subsequent microscope images that cover the full device area. It accounts for problems that generally occur in fluorescent lab-chip cell images such as: uneven background, overlapping cell images and cell detection with multiple stains. This method can be used in laboratories to save time and effort, and to increase cell counting accuracy of lab-chip devices for various applications, such as circulating tumor cell detection, cell detection in biosensors, and HIV monitoring devices, i.e. CD4 counts.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

