Structural differences between apolipoprotein E3 and E4 as measured by (19)F NMR
- PMID: 19904741
- PMCID: PMC2817840
- DOI: 10.1002/pro.283
Structural differences between apolipoprotein E3 and E4 as measured by (19)F NMR
Abstract
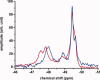
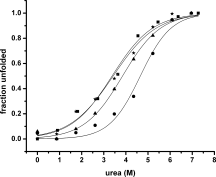
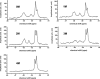
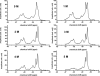
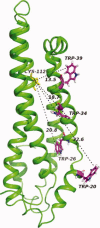
The apolipoprotein E family contains three major isoforms (ApoE4, E3, and E2) that are directly involved with lipoprotein metabolism and cholesterol transport. ApoE3 and apoE4 differ in only a single amino acid with an arginine in apoE4 changed to a cysteine at position 112 in apoE3. Yet only apoE4 is recognized as a risk factor for Alzheimer's disease. Here we used (19)F NMR to examine structural differences between apoE4 and apoE3 and the effect of the C-terminal domain on the N-terminal domain. After incorporation of 5-(19)F-tryptophan the 1D (19)F NMR spectra were compared for the N-terminal domain and for the full length proteins. The NMR spectra of the N-terminal region (residues 1-191) are reasonably well resolved while those of the full length wild-type proteins are broad and ill-defined suggesting considerable conformational heterogeneity. At least four of the seven tryptophan residues in the wild type protein appear to be solvent exposed. NMR spectra of the wild-type proteins were compared to apoE containing four mutations in the C-terminal region that gives rise to a monomeric form either of apoE3 under native conditions (Zhang et al., Biochemistry 2007; 46: 10722-10732) or apoE4 in the presence of 1 M urea. For either wild-type or mutant proteins the differences in tryptophan resonances in the N-terminal region of the protein suggest structural differences between apoE3 and apoE4. We conclude that these differences occur both as a consequence of the Arg158Cys mutation and as a consequence of the interaction with the C-terminal domain.
Figures


References
-
- Aggerbeck LP, Wetterau JR, Weisgraber KH, Wu CS, Lindgren FT. Human apolipoprotein E3 in aqueous solution. II. Properties of the amino- and carboxyl-terminal domains. J Biol Chem. 1988;263:6249–6258. - PubMed
-
- Hatters DM, Peters-Libeu CA, Weisgraber KH. Apolipoprotein E structure: insights into function. Trends Biochem Sci. 2006;31:445–454. - PubMed
-
- Perugini MA, Schuck P, Howlett GJ. Self-association of human apolipoprotein E3 and E4 in the presence and absence of phospholipid. J Biol Chem. 2000;275:36758–36765. - PubMed
-
- Yokoyama S, Kawai Y, Tajima S, Yamamoto A. Behavior of human apolipoprotein E in aqueous solutions and at interfaces. J Biol Chem. 1985;260:16375–16382. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

