The immune epitope database 2.0
- PMID: 19906713
- PMCID: PMC2808938
- DOI: 10.1093/nar/gkp1004
The immune epitope database 2.0
Abstract
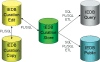
The Immune Epitope Database (IEDB, www.iedb.org) provides a catalog of experimentally characterized B and T cell epitopes, as well as data on Major Histocompatibility Complex (MHC) binding and MHC ligand elution experiments. The database represents the molecular structures recognized by adaptive immune receptors and the experimental contexts in which these molecules were determined to be immune epitopes. Epitopes recognized in humans, nonhuman primates, rodents, pigs, cats and all other tested species are included. Both positive and negative experimental results are captured. Over the course of 4 years, the data from 180,978 experiments were curated manually from the literature, which covers approximately 99% of all publicly available information on peptide epitopes mapped in infectious agents (excluding HIV) and 93% of those mapped in allergens. In addition, data that would otherwise be unavailable to the public from 129,186 experiments were submitted directly by investigators. The curation of epitopes related to autoimmunity is expected to be completed by the end of 2010. The database can be queried by epitope structure, source organism, MHC restriction, assay type or host organism, among other criteria. The database structure, as well as its querying, browsing and reporting interfaces, was completely redesigned for the IEDB 2.0 release, which became publicly available in early 2009.
Figures


References
-
- Blythe MJ, Doytchinova IA, Flower DR. JenPep: a database of quantitative functional peptide data for immunology. Bioinformatics. 2002;18:434–439. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

