Molecular mechanisms of RNA polymerase--the F/E (RPB4/7) complex is required for high processivity in vitro
- PMID: 19906731
- PMCID: PMC2811020
- DOI: 10.1093/nar/gkp928
Molecular mechanisms of RNA polymerase--the F/E (RPB4/7) complex is required for high processivity in vitro
Abstract
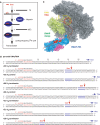
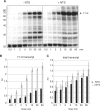
Transcription elongation in vitro is affected by the interactions between RNA polymerase (RNAP) subunits and the nucleic acid scaffold of the ternary elongation complex (TEC, RNAP-DNA-RNA). We have investigated the role of the RNAP subunits F/E (homologous to eukaryotic RPB4/7) during transcription elongation and termination using a wholly recombinant archaeal RNAP and synthetic nucleic acid scaffolds. The F/E complex greatly stimulates the processivity of RNAP, it enhances the formation of full length products, reduces pausing, and increases transcription termination facilitated by weak termination signals. Mutant variants of F/E that are defective in RNA binding show that these activities correlate with the nucleic acid binding properties of F/E. However, a second RNA-binding independent component also contributes to the stimulatory activities of F/E. In summary, our results suggest that interactions between RNAP subunits F/E and the RNA transcript are pivotal to the molecular mechanisms of RNAP during transcription elongation and termination.
Figures

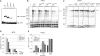

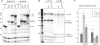
Similar articles
-
RNAP subunits F/E (RPB4/7) are stably associated with archaeal RNA polymerase: using fluorescence anisotropy to monitor RNAP assembly in vitro.Biochem J. 2009 Jul 15;421(3):339-43. doi: 10.1042/BJ20090782. Biochem J. 2009. PMID: 19492989
-
Hold on!: RNA polymerase interactions with the nascent RNA modulate transcription elongation and termination.RNA Biol. 2010 May-Jun;7(3):310-5. doi: 10.4161/rna.7.3.11912. Epub 2010 May 26. RNA Biol. 2010. PMID: 20473037 Free PMC article. Review.
-
Cycling through transcription with the RNA polymerase F/E (RPB4/7) complex: structure, function and evolution of archaeal RNA polymerase.Res Microbiol. 2011 Jan;162(1):10-8. doi: 10.1016/j.resmic.2010.09.002. Epub 2010 Sep 21. Res Microbiol. 2011. PMID: 20863887 Review.
-
Evolution of complex RNA polymerases: the complete archaeal RNA polymerase structure.PLoS Biol. 2009 May;7(5):e1000102. doi: 10.1371/journal.pbio.1000102. Epub 2009 May 5. PLoS Biol. 2009. PMID: 19419240 Free PMC article.
-
Archaeal RNA polymerase subunits E and F are not required for transcription in vitro, but a Thermococcus kodakarensis mutant lacking subunit F is temperature-sensitive.Mol Microbiol. 2008 Nov;70(3):623-33. doi: 10.1111/j.1365-2958.2008.06430.x. Epub 2008 Sep 10. Mol Microbiol. 2008. PMID: 18786148 Free PMC article.
Cited by
-
Structure of the recombinant RNA polymerase from African Swine Fever Virus.Nat Commun. 2024 Feb 21;15(1):1606. doi: 10.1038/s41467-024-45842-7. Nat Commun. 2024. PMID: 38383525 Free PMC article.
-
RNA polymerase backtracking in gene regulation and genome instability.Cell. 2012 Jun 22;149(7):1438-45. doi: 10.1016/j.cell.2012.06.003. Cell. 2012. PMID: 22726433 Free PMC article.
-
aCPSF1 cooperates with terminator U-tract to dictate archaeal transcription termination efficacy.Elife. 2021 Dec 29;10:e70464. doi: 10.7554/eLife.70464. Elife. 2021. PMID: 34964713 Free PMC article.
-
Molecular Mechanisms of Transcription Initiation-Structure, Function, and Evolution of TFE/TFIIE-Like Factors and Open Complex Formation.J Mol Biol. 2016 Jun 19;428(12):2592-2606. doi: 10.1016/j.jmb.2016.04.016. Epub 2016 Apr 20. J Mol Biol. 2016. PMID: 27107643 Free PMC article. Review.
-
The RNA polymerase trigger loop functions in all three phases of the transcription cycle.Nucleic Acids Res. 2013 Aug;41(14):7048-59. doi: 10.1093/nar/gkt433. Epub 2013 Jun 3. Nucleic Acids Res. 2013. PMID: 23737452 Free PMC article.
References
-
- Werner F. Structural evolution of multisubunit RNA polymerases. Trends Microbiol. 2008;16:247–250. - PubMed
-
- Werner F. Structure and function of archaeal RNA polymerases. Mol. Microbiol. 2007;65:1395–1404. - PubMed
-
- Grohmann D, Hirtreiter A, Werner F. The RNAP subunits F/E (RPB4/7) are stably associated with archaeal RNA polymerase – using fluorescence anisotropy to monitor RNAP assembly in vitro. Biochem. J. 2009;421:339–343. - PubMed
-
- Ujvari A, Luse DS. RNA emerging from the active site of RNA polymerase II interacts with the Rpb7 subunit. Nat. Struct. Mol. Biol. 2006;13:49–54. - PubMed

