Sanpodo: a context-dependent activator and inhibitor of Notch signaling during asymmetric divisions
- PMID: 19906847
- PMCID: PMC2781049
- DOI: 10.1242/dev.040386
Sanpodo: a context-dependent activator and inhibitor of Notch signaling during asymmetric divisions
Abstract
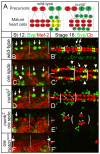
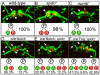
Asymmetric cell divisions generate sibling cells of distinct fates ('A', 'B') and constitute a fundamental mechanism that creates cell-type diversity in multicellular organisms. Antagonistic interactions between the Notch pathway and the intrinsic cell-fate determinant Numb appear to regulate asymmetric divisions in flies and vertebrates. During these divisions, productive Notch signaling requires sanpodo, which encodes a novel transmembrane protein. Here, we demonstrate that Drosophila sanpodo plays a dual role to regulate Notch signaling during asymmetric divisions - amplifying Notch signaling in the absence of Numb in the 'A' daughter cell and inhibiting Notch signaling in the presence of Numb in the 'B' daughter cell. In so doing, sanpodo ensures the asymmetry in Notch signaling levels necessary for the acquisition of distinct fates by the two daughter cells. These findings answer long-standing questions about the restricted ability of Numb and Sanpodo to inhibit and to promote, respectively, Notch signaling during asymmetric divisions.
Figures






References
-
- Ahmed A., Chandra S., Magarinos M., Vaessin H. (2003). Echinoid mutants exhibit neurogenic phenotypes and show synergistic interactions with the Notch signaling pathway. Development 130, 6295-6304 - PubMed
-
- Berdnik D., Torok T., Gonzalez-Gaitan M., Knoblich J. A. (2002). The endocytic protein alpha-Adaptin is required for numb-mediated asymmetric cell division in Drosophila. Dev. Cell 3, 221-231 - PubMed
-
- Blair S. S. (2000). Notch signaling: Fringe really is a glycosyltransferase. Curr. Biol. 10, R608-R612 - PubMed
-
- Brand A. H., Perrimon N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401-415 - PubMed
-
- Calleja M., Moreno E., Pelaz S., Morata G. (1996). Visualization of gene expression in living adult Drosophila. Science 274, 252-255 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

