Large-scale reverse genetics in Arabidopsis: case studies from the Chloroplast 2010 Project
- PMID: 19906890
- PMCID: PMC2815874
- DOI: 10.1104/pp.109.148494
Large-scale reverse genetics in Arabidopsis: case studies from the Chloroplast 2010 Project
Abstract
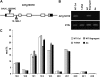
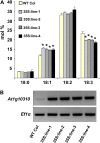
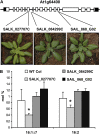
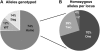
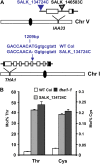
Traditionally, phenotype-driven forward genetic plant mutant studies have been among the most successful approaches to revealing the roles of genes and their products and elucidating biochemical, developmental, and signaling pathways. A limitation is that it is time consuming, and sometimes technically challenging, to discover the gene responsible for a phenotype by map-based cloning or discovery of the insertion element. Reverse genetics is also an excellent way to associate genes with phenotypes, although an absence of detectable phenotypes often results when screening a small number of mutants with a limited range of phenotypic assays. The Arabidopsis Chloroplast 2010 Project (www.plastid.msu.edu) seeks synergy between forward and reverse genetics by screening thousands of sequence-indexed Arabidopsis (Arabidopsis thaliana) T-DNA insertion mutants for a diverse set of phenotypes. Results from this project are discussed that highlight the strengths and limitations of the approach. We describe the discovery of altered fatty acid desaturation phenotypes associated with mutants of At1g10310, previously described as a pterin aldehyde reductase in folate metabolism. Data are presented to show that growth, fatty acid, and chlorophyll fluorescence defects previously associated with antisense inhibition of synthesis of the family of acyl carrier proteins can be attributed to a single gene insertion in Acyl Carrier Protein4 (At4g25050). A variety of cautionary examples associated with the use of sequence-indexed T-DNA mutants are described, including the need to genotype all lines chosen for analysis (even when they number in the thousands) and the presence of tagged and untagged secondary mutations that can lead to the observed phenotypes.
Figures


References
-
- Adham AR, Zolman BK, Millius A, Bartel B (2005) Mutations in Arabidopsis acyl-CoA oxidase genes reveal distinct and overlapping roles in beta-oxidation. Plant J 41 859–874 - PubMed
-
- Ajjawi I, Rodriguez Milla MA, Cushman J, Shintani DK (2007) Thiamin pyrophosphokinase is required for thiamin cofactor activation in Arabidopsis. Plant Mol Biol 65 151–162 - PubMed
-
- Alonso JM, Ecker JR (2006) Moving forward in reverse: genetic technologies to enable genome-wide phenomic screens in Arabidopsis. Nat Rev Genet 7 524–536 - PubMed
-
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

