Identification of GBF1 as a cellular factor required for hepatitis C virus RNA replication
- PMID: 19906930
- PMCID: PMC2798365
- DOI: 10.1128/JVI.01190-09
Identification of GBF1 as a cellular factor required for hepatitis C virus RNA replication
Abstract
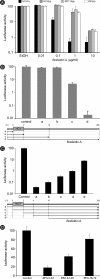
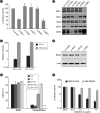
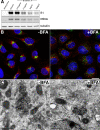
In infected cells, hepatitis C virus (HCV) induces the formation of membrane alterations referred to as membranous webs, which are sites of RNA replication. In addition, HCV RNA replication also occurs in smaller membrane structures that are associated with the endoplasmic reticulum. However, cellular mechanisms involved in the formation of HCV replication complexes remain largely unknown. Here, we used brefeldin A (BFA) to investigate cellular mechanisms involved in HCV infection. BFA acts on cell membranes by interfering with the activation of several members of the family of ADP-ribosylation factors (ARF), which can lead to a wide range of inhibitory actions on membrane-associated mechanisms of the secretory and endocytic pathways. Our data show that HCV RNA replication is highly sensitive to BFA. Individual knockdown of the cellular targets of BFA using RNA interference and the use of a specific pharmacological inhibitor identified GBF1, a guanine nucleotide exchange factor for small GTPases of the ARF family, as a host factor critically involved in HCV replication. Furthermore, overexpression of a BFA-resistant GBF1 mutant rescued HCV replication in BFA-treated cells, indicating that GBF1 is the BFA-sensitive factor required for HCV replication. Finally, immunofluorescence and electron microscopy analyses indicated that BFA does not block the formation of membranous web-like structures induced by expression of HCV proteins in a nonreplicative context, suggesting that GBF1 is probably involved not in the formation of HCV replication complexes but, rather, in their activity. Altogether, our results highlight a functional connection between the early secretory pathway and HCV RNA replication.
Figures





References
-
- Belouzard, S., D. Delcroix, and Y. Rouillé. 2004. Low levels of expression of leptin receptor at the cell surface result from constitutive endocytosis and intracellular retention in the biosynthetic pathway. J. Biol. Chem. 279:28499-28508. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

