Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function
- PMID: 19906987
- PMCID: PMC2775998
- DOI: 10.1073/pnas.0908365106
Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function
Abstract
HBV cccDNA, the template for transcription of all viral mRNAs, accumulates in the nucleus of infected cells as a stable episome organized into minichromosomes by histones and non-histone viral and cellular proteins. Using a cccDNA-specific chromatin immunoprecipitation (ChIP)-based quantitative assay, we have previously shown that transcription of the HBV minichromosome is regulated by epigenetic changes of cccDNA-bound histones and that modulation of the acetylation status of cccDNA-bound H3/H4 histones impacts on HBV replication. We now show that the cellular histone acetyltransferases CBP, p300, and PCAF/GCN5, and the histone deacetylases HDAC1 and hSirt1 are all recruited in vivo onto the cccDNA. We also found that the HBx regulatory protein produced in HBV replicating cells is recruited onto the cccDNA minichromosome, and the kinetics of HBx recruitment on the cccDNA parallels the HBV replication. As expected, an HBV mutant that does not express HBx is impaired in its replication, and exogenously expressed HBx transcomplements the replication defects. p300 recruitment is severely impaired, and cccDNA-bound histones are rapidly hypoacetylated in cells replicating the HBx mutant, whereas the recruitment of the histone deacetylases hSirt1 and HDAC1 is increased and occurs at earlier times. Finally, HBx mutant cccDNA transcribes significantly less pgRNA. Altogether our results further support the existence of a complex network of epigenetic events that influence cccDNA function and HBV replication and identify an epigenetic mechanism (i.e., to prevent cccDNA deacetylation) by which HBx controls HBV replication.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


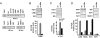
References
-
- Shepard CW, Simard EP, Finelli L, Fiore AE, Bell BP. Hepatitis B virus infection: Epidemiology and vaccination. Epidemiol Rev. 2006;28:112–125. - PubMed
-
- Beasley RP. Hepatitis B virus. The major etiology of hepatocellular carcinoma. Cancer. 1988;61:1942–1956. - PubMed
-
- Buendia MA. Genetics of hepatocellular carcinoma. Semin Cancer Biol. 2000;10:185–200. - PubMed
-
- Kremsdorf D, Soussan P, Paterlini-Brechot P, Brechot C. Hepatitis B virus-related hepatocellular carcinoma: Paradigms for viral-related human carcinogenesis. Oncogene. 2006;25:3823–3833. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

