Human BLCAP transcript: new editing events in normal and cancerous tissues
- PMID: 19908260
- PMCID: PMC2958456
- DOI: 10.1002/ijc.25022
Human BLCAP transcript: new editing events in normal and cancerous tissues
Abstract
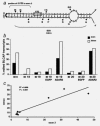
Bladder cancer-associated protein (BLCAP) is a highly conserved protein among species, and it is considered a novel candidate tumor suppressor gene originally identified from human bladder carcinoma. However, little is known about the regulation or the function of this protein. Here, we show that the human BLCAP transcript undergoes multiple A-to-I editing events. Some of the new editing events alter the highly conserved amino terminus of the protein creating alternative protein isoforms by changing the genetically coded amino acids. We found that both ADAR1 and ADAR2-editing enzymes cooperate to edit this transcript and that different tissues displayed distinctive ratios of edited and unedited BLCAP transcripts. Moreover, we observed a general decrease in BLCAP-editing level in astrocytomas, bladder cancer and colorectal cancer when compared with the related normal tissues. The newly identified editing events, found to be downregulated in cancers, could be useful for future studies as a diagnostic tool to distinguish malignancies or epigenetic changes in different tumors.
Figures


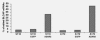

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

