Predicting RNA structure by multiple template homology modeling
- PMID: 19908374
- PMCID: PMC2872935
- DOI: 10.1142/9789814295291_0024
Predicting RNA structure by multiple template homology modeling
Abstract
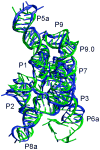
Despite the importance of 3D structure to understand the myriad functions of RNAs in cells, most RNA molecules remain out of reach of crystallographic and NMR methods. However, certain structural information such as base pairing and some tertiary contacts can be determined readily for many RNAs by bioinformatics or relatively low cost experiments. Further, because RNA structure is highly modular, it is possible to deduce local 3D structure from the solved structures of evolutionarily related RNAs or even unrelated RNAs that share the same module. RNABuilder is a software package that generates model RNA structures by treating the kinematics and forces at separate, multiple levels of resolution. Kinematically, bonds in bases, certain stretches of residues, and some entire molecules are rigid while other bonds remain flexible. Forces act on the rigid bases and selected individual atoms. Here we use RNABuilder to predict the structure of the 200-nucleotide Azoarcus group I intron by homology modeling against fragments of the distantly-related Twort and Tetrahymena group I introns and by incorporating base pairing forces where necessary. In the absence of any information from the solved Azoarcus intron crystal structure, the model accurately depicts the global topology, secondary and tertiary connections, and gives an overall RMSD value of 4.6 A relative to the crystal structure. The accuracy of the model is even higher in the intron core (RMSD = 3.5 A), whereas deviations are modestly larger for peripheral regions that differ more substantially between the different introns. These results lay the groundwork for using this approach for larger and more diverse group I introns, as well for still larger RNAs and RNA-protein complexes such as group II introns and the ribosomal subunits.
Figures